Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae
- PMID: 20398375
- PMCID: PMC3095342
- DOI: 10.1186/1471-2229-10-68
Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae
Abstract
Background: Oncidium spp. produce commercially important orchid cut flowers. However, they are amenable to intergeneric and inter-specific crossing making phylogenetic identification very difficult. Molecular markers derived from the chloroplast genome can provide useful tools for phylogenetic resolution.
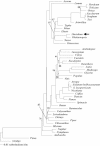
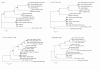
Results: The complete chloroplast genome of the economically important Oncidium variety Onc. Gower Ramsey (Accession no. GQ324949) was determined using a polymerase chain reaction (PCR) and Sanger based ABI sequencing. The length of the Oncidium chloroplast genome is 146,484 bp. Genome structure, gene order and orientation are similar to Phalaenopsis, but differ from typical Poaceae, other monocots for which there are several published chloroplast (cp) genome. The Onc. Gower Ramsey chloroplast-encoded NADH dehydrogenase (ndh) genes, except ndhE, lack apparent functions. Deletion and other types of mutations were also found in the ndh genes of 15 other economically important Oncidiinae varieties, except ndhE in some species. The positions of some species in the evolution and taxonomy of Oncidiinae are difficult to identify. To identify the relationships between the 15 Oncidiinae hybrids, eight regions of the Onc. Gower Ramsey chloroplast genome were amplified by PCR for phylogenetic analysis. A total of 7042 bp derived from the eight regions could identify the relationships at the species level, which were supported by high bootstrap values. One particular 1846 bp region, derived from two PCR products (trnHGUG -psbA and trnFGAA-ndhJ) was adequate for correct phylogenetic placement of 13 of the 15 varieties (with the exception of Degarmoara Flying High and Odontoglossum Violetta von Holm). Thus the chloroplast genome provides a useful molecular marker for species identifications.
Conclusion: In this report, we used Phalaenopsis. aphrodite as a prototype for primer design to complete the Onc. Gower Ramsey genome sequence. Gene annotation showed that most of the ndh genes inOncidiinae, with the exception of ndhE, are non-functional. This phenomenon was observed in all of the Oncidiinae species tested. The genes and chloroplast DNA regions that would be the most useful for phylogenetic analysis were determined to be the trnHGUG-psbA and the trnFGAA-ndhJ regions. We conclude that complete chloroplast genome information is useful for plant phylogenetic and evolutionary studies in Oncidium with applications for breeding and variety identification.
Figures





References
-
- Webster P. The Orchid Genus Book. Hunters Breeze, USA; 1992. Oncidium subtribe.
-
- Chase W, Palmer JD. Chloroplast DNA systematics of lilioid monocots: resources, feasibility, and an example from the Orchidaceae. Amer J Bot. 1989;76:1720–1730. doi: 10.2307/2444471. - DOI
-
- Tsai CC, Huang SC, Huang PL, Chen YS, Chou CH. Phenetic relationship and identification of subtribe Oncidiinae genotypes by random amplified polymorphic DNA (RAPD) markers. Sci Hort. 2002;101:315–325. doi: 10.1016/j.scienta.2003.11.004. - DOI
-
- Tien X, Li DZ. Application of DNA sequences in plant phylogenetic study. Acta Bot Yumnan. 2002;24:170–184.
-
- Cao Q-q, Zhen Z, Jiang J, Liu Y-f, Feng Y-q, Qin L. Chloroplast DNA analysis technology and its application in Castanea. J Fruit Sci. 2008;25:396–399.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

