The extended PP1 toolkit: designed to create specificity
- PMID: 20399103
- PMCID: PMC3131691
- DOI: 10.1016/j.tibs.2010.03.002
The extended PP1 toolkit: designed to create specificity
Abstract
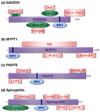
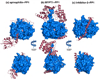
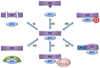
Protein Ser/Thr phosphatase-1 (PP1) catalyzes the majority of eukaryotic protein dephosphorylation reactions in a highly regulated and selective manner. Recent studies have identified an unusually diversified PP1 interactome with the properties of a regulatory toolkit. PP1-interacting proteins (PIPs) function as targeting subunits, substrates and/or inhibitors. As targeting subunits, PIPs contribute to substrate selection by bringing PP1 into the vicinity of specific substrates and by modulating substrate specificity via additional substrate docking sites or blocking substrate-binding channels. Many of the nearly 200 established mammalian PIPs are predicted to be intrinsically disordered, a property that facilitates their binding to a large surface area of PP1 via multiple docking motifs. These novel insights offer perspectives for the therapeutic targeting of PP1 by interfering with the binding of PIPs or substrates.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

References
-
- Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. - PubMed
-
- Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. - PubMed
-
- Manning G, et al. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002;27:514–520. - PubMed
-
- Moorhead GB, et al. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 2007;8:234–244. - PubMed
-
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009;33:537–545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

