The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma
- PMID: 20399120
- PMCID: PMC2867662
- DOI: 10.1016/j.immuni.2010.04.004
The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma
Abstract
The transcription factor GATA3 is crucial for the differentiation of naive CD4(+) T cells into T helper 2 (Th2) cells. Here, we show that deletion of Gata3 allowed the appearance of interferon-gamma (IFN-gamma)-producing cells in the absence of interleukin-12 (IL-12) and IFN-gamma. Such IFN-gamma production was transcription factor T-bet independent. Another T-box-containing transcription factor Eomes, but not T-bet, was induced both in GATA3-deficient CD4(+) T cells differentiated under Th2 cell conditions and in Th2 cells with enforced Runx3 expression, contributing to IFN-gamma production. GATA3 overexpression blocked Runx3-mediated Eomes induction and IFN-gamma production, and GATA3 protein physically interacted with Runx3 protein. Furthermore, we found that Runx3 directly bound to multiple regulatory elements of the Ifng gene and that blocking Runx3 function in either Th1 or GATA3-deficient "Th2" cells results in diminished IFN-gamma production by these cells. Thus, the Runx3-mediated pathway, actively suppressed by GATA3, induces IFN-gamma production in a STAT4- and T-bet-independent manner.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


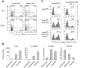

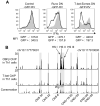

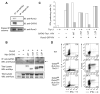
Comment in
-
Antisocial networking in T helper cells.Immunity. 2010 Apr 23;32(4):500-1. doi: 10.1016/j.immuni.2010.04.006. Immunity. 2010. PMID: 20412760
References
-
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nature immunology. 2002;3:549–557. - PubMed
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bosselut R. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol. 2004;4:529–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

