The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems
- PMID: 20400551
- PMCID: PMC2916427
- DOI: 10.1128/JB.00201-10
The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems
Erratum in
- J Bacteriol. 2011 Jan;193(1):326
Abstract
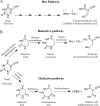
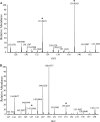
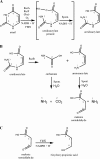
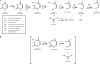
The Rut pathway is composed of seven proteins, all of which are required by Escherichia coli K-12 to grow on uracil as the sole nitrogen source. The RutA and RutB proteins are central: no spontaneous suppressors arise in strains lacking them. RutA works in conjunction with a flavin reductase (RutF or a substitute) to catalyze a novel reaction. It directly cleaves the uracil ring between N-3 and C-4 to yield ureidoacrylate, as established by both nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry. Although ureidoacrylate appears to arise by hydrolysis, the requirements for the reaction and the incorporation of (18)O at C-4 from molecular oxygen indicate otherwise. Mass spectrometry revealed the presence of a small amount of product with the mass of ureidoacrylate peracid in reaction mixtures, and we infer that this is the direct product of RutA. In vitro RutB cleaves ureidoacrylate hydrolytically to release 2 mol of ammonium, malonic semialdehyde, and carbon dioxide. Presumably the direct products are aminoacrylate and carbamate, both of which hydrolyze spontaneously. Together with bioinformatic predictions and published crystal structures, genetic and physiological studies allow us to predict functions for RutC, -D, and -E. In vivo we postulate that RutB hydrolyzes the peracid of ureidoacrylate to yield the peracid of aminoacrylate. We speculate that RutC reduces aminoacrylate peracid to aminoacrylate and RutD increases the rate of spontaneous hydrolysis of aminoacrylate. The function of RutE appears to be the same as that of YdfG, which reduces malonic semialdehyde to 3-hydroxypropionic acid. RutG appears to be a uracil transporter.
Figures


Comment in
-
The surprising Rut pathway: an unexpected way to derive nitrogen from pyrimidines.J Bacteriol. 2010 Aug;192(16):4086-8. doi: 10.1128/JB.00573-10. Epub 2010 Jun 18. J Bacteriol. 2010. PMID: 20562306 Free PMC article. No abstract available.
References
-
- Andersen, G., O. Björnberg, S. Polakova, Y. Pynyaha, A. Rasmussen, K. Møller, A. Hofer, T. Moritz, M. P. Sandrini, A. M. Merico, C. Compagno, H. E. Akerlund, Z. Gojković, and J. Piskur. 2008. A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J. Mol. Biol. 380:655-666. - PubMed
-
- Andersen, K. B., and K. von Meyenburg. 1977. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J. Biol. Chem. 252:4151-4156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

