Cleavage site specificity of MMP-20 for secretory-stage ameloblastin
- PMID: 20400724
- PMCID: PMC2909333
- DOI: 10.1177/0022034510366903
Cleavage site specificity of MMP-20 for secretory-stage ameloblastin
Abstract
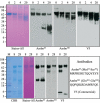
Ameloblastin is processed by protease(s) during enamel formation. We tested the hypothesis that MMP-20 (enamelysin) catalyzes the cleavages that generate secretory-stage ameloblastin cleavage products. We isolated a 23-kDa ameloblastin cleavage product from developing enamel and determined its N-terminus sequence. Ameloblastin was stably expressed and secreted from HEK293-H cells, purified, and digested with MMP-20 or Klk4 (kallikrein 4). The digests were analyzed by SDS-PAGE and Western blotting, and cleavage products were characterized by N-terminal sequencing. Six fluorescent peptides were digested with MMP-20 and Klk4 and analyzed by RP-HPLC and by mass spectrometry. MMP-20 cleaved each peptide exactly at the sites corresponding to ameloblastin cleavages catalyzed in vivo. Klk4 cleaved ameloblastin and the fluorescent peptides at sites not observed in vivo, and cleaved at only a single correct site: before Leu(171). We conclude that MMP-20 is the enzyme that processes ameloblastin during the secretory stage of amelogenesis, and we present a hypothesis about the sequence of ameloblastin cleavages.
Conflict of interest statement
All authors declare that there are no conflicting interests.
Figures



References
-
- Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. (1996). Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 183:123-128 - PubMed
-
- Bartlett JD, Ryu OH, Xue J, Simmer JP, Margolis HC. (1998). Enamelysin mRNA displays a developmentally defined pattern of expression and encodes a protein which degrades amelogenin. Connect Tissue Res 39:101-109 - PubMed
-
- Beyeler M, Schild C, Lutz R, Chiquet M, Trueb B. (2010). Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp Cell Res [Epub ahead of print], January 4, 2010 (in press) - PubMed
-
- Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, et al. (2002). Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 277:49598-49604 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

