Replication through an abasic DNA lesion: structural basis for adenine selectivity
- PMID: 20400942
- PMCID: PMC2876968
- DOI: 10.1038/emboj.2010.64
Replication through an abasic DNA lesion: structural basis for adenine selectivity
Abstract
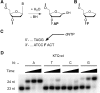
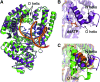
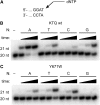
Abasic sites represent the most frequent DNA lesions in the genome that have high mutagenic potential and lead to mutations commonly found in human cancers. Although these lesions are devoid of the genetic information, adenine is most efficiently inserted when abasic sites are bypassed by DNA polymerases, a phenomenon termed A-rule. In this study, we present X-ray structures of a DNA polymerase caught while incorporating a nucleotide opposite an abasic site. We found that a functionally important tyrosine side chain directs for nucleotide incorporation rather than DNA. It fills the vacant space of the absent template nucleobase and thereby mimics a pyrimidine nucleobase directing for preferential purine incorporation opposite abasic residues because of enhanced geometric fit to the active site. This amino acid templating mechanism was corroborated by switching to pyrimidine specificity because of mutation of the templating tyrosine into tryptophan. The tyrosine is located in motif B and highly conserved throughout evolution from bacteria to humans indicating a general amino acid templating mechanism for bypass of non-instructive lesions by DNA polymerases at least from this sequence family.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

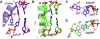

Similar articles
-
Amino acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.J Biol Chem. 2012 Apr 20;287(17):14099-108. doi: 10.1074/jbc.M111.334904. Epub 2012 Feb 7. J Biol Chem. 2012. PMID: 22318723 Free PMC article.
-
New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions.Mutat Res. 2002 Dec 29;510(1-2):55-70. doi: 10.1016/s0027-5107(02)00252-x. Mutat Res. 2002. PMID: 12459443 Review.
-
Structural and kinetic analysis of nucleoside triphosphate incorporation opposite an abasic site by human translesion DNA polymerase η.J Biol Chem. 2015 Mar 27;290(13):8028-38. doi: 10.1074/jbc.M115.637561. Epub 2015 Feb 9. J Biol Chem. 2015. PMID: 25666608 Free PMC article.
-
Pyrimidine ring fragmentation products. Effects of lesion structure and sequence context on mutagenesis.J Mol Biol. 1994 Feb 18;236(2):514-30. doi: 10.1006/jmbi.1994.1162. J Mol Biol. 1994. PMID: 8107137
-
Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass.DNA Repair (Amst). 2002 May 30;1(5):343-58. doi: 10.1016/s1568-7864(02)00019-8. DNA Repair (Amst). 2002. PMID: 12509239 Review.
Cited by
-
Base-pairing preferences, physicochemical properties and mutational behaviour of the DNA lesion 8-nitroguanine.Nucleic Acids Res. 2012 Nov;40(21):11126-38. doi: 10.1093/nar/gks799. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965127 Free PMC article.
-
The Structural Basis for Processing of Unnatural Base Pairs by DNA Polymerases.Chemistry. 2020 Mar 18;26(16):3446-3463. doi: 10.1002/chem.201903525. Epub 2020 Jan 21. Chemistry. 2020. PMID: 31544987 Free PMC article. Review.
-
The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site.Biochemistry. 2011 Nov 29;50(47):10350-8. doi: 10.1021/bi201219s. Epub 2011 Nov 3. Biochemistry. 2011. PMID: 22026756 Free PMC article.
-
Identification of Thermus aquaticus DNA polymerase variants with increased mismatch discrimination and reverse transcriptase activity from a smart enzyme mutant library.Sci Rep. 2019 Jan 24;9(1):590. doi: 10.1038/s41598-018-37233-y. Sci Rep. 2019. PMID: 30679705 Free PMC article.
-
Synthetic nucleotides as probes of DNA polymerase specificity.J Nucleic Acids. 2012;2012:530963. doi: 10.1155/2012/530963. Epub 2012 Jun 7. J Nucleic Acids. 2012. PMID: 22720133 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D 58: 1948–1954 - PubMed
-
- Bell JB, Eckert KA, Joyce CM, Kunkel TA (1997) Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J Biol Chem 272: 7345–7351 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

