Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1
- PMID: 20400958
- PMCID: PMC2866742
- DOI: 10.1038/ncb2049
Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1
Abstract
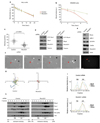
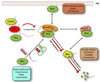
At steady state, most Rho GTPases are bound in the cytosol to Rho guanine nucleotide dissociation inhibitors (RhoGDIs). RhoGDIs have generally been considered to hold Rho proteins passively in an inactive state within the cytoplasm. Here we describe an evolutionarily conserved mechanism by which RhoGDI1 controls the homeostasis of Rho proteins in eukaryotic cells. We found that depletion of RhoGDI1 promotes misfolding and degradation of the cytosolic geranylgeranylated pool of Rho GTPases while activating the remaining membrane-bound fraction. Because RhoGDI1 levels are limiting, and Rho proteins compete for binding to RhoGDI1, overexpression of an exogenous Rho GTPase displaces endogenous Rho proteins bound to RhoGDI1, inducing their degradation and inactivation. These results raise important questions about the conclusions drawn from studies that manipulate Rho protein levels. In many cases the response observed may arise not simply from the overexpression itself but from additional effects on the levels and activity of other Rho GTPases as a result of competition for binding to RhoGDI1; this may require a re-evaluation of previously published studies that rely exclusively on these techniques.
Figures



References
-
- DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. - PubMed
-
- Dransart E, Morin A, Cherfils J, Olofsson B. Uncoupling of inhibitory and shuttling functions of rho GDP dissociation inhibitors. J Biol Chem. 2005;280:4674–4683. - PubMed
-
- Gorovoy M, et al. RhoGDI-1 modulation of the activity of monomeric RhoGTPase RhoA regulates endothelial barrier function in mouse lungs. Circ Res. 2007;101:50–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

