The kinetics of antibody binding to Plasmodium falciparum VAR2CSA PfEMP1 antigen and modelling of PfEMP1 antigen packing on the membrane knobs
- PMID: 20403153
- PMCID: PMC2868858
- DOI: 10.1186/1475-2875-9-100
The kinetics of antibody binding to Plasmodium falciparum VAR2CSA PfEMP1 antigen and modelling of PfEMP1 antigen packing on the membrane knobs
Abstract
Background: Infected humans make protective antibody responses to the PfEMP1 adhesion antigens exported by Plasmodium falciparum parasites to the erythrocyte membrane, but little is known about the kinetics of this antibody-receptor binding reaction or how the topology of PfEMP1 on the parasitized erythrocyte membrane influences antibody association with, and dissociation from, its antigenic target.
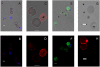
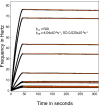
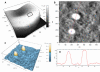
Methods: A Quartz Crystal Microbalance biosensor was used to measure the association and dissociation kinetics of VAR2CSA PfEMP1 binding to human monoclonal antibodies. Immuno-fluorescence microscopy was used to visualize antibody-mediated adhesion between the surfaces of live infected erythrocytes and atomic force microscopy was used to obtain higher resolution images of the membrane knobs on the infected erythrocyte to estimate knob surface areas and model VAR2CSA packing density on the knob.
Results: Kinetic analysis indicates that antibody dissociation from the VAR2CSA PfEMP1 antigen is extremely slow when there is a high avidity interaction. High avidity binding to PfEMP1 antigens on the surface of P. falciparum-infected erythrocytes in turn requires bivalent cross-linking of epitopes positioned within the distance that can be bridged by antibody. Calculations of the surface area of the knobs and the possible densities of PfEMP1 packing on the knobs indicate that high-avidity cross-linking antibody reactions are constrained by the architecture of the knobs and the large size of PfEMP1 molecules.
Conclusions: High avidity is required to achieve the strongest binding to VAR2CSA PfEMP1, but the structures that display PfEMP1 also tend to inhibit cross-linking between PfEMP1 antigens, by holding many binding epitopes at distances beyond the 15-18 nm sweep radius of an antibody. The large size of PfEMP1 will also constrain intra-knob cross-linking interactions. This analysis indicates that effective vaccines targeting the parasite's vulnerable adhesion receptors should primarily induce strongly adhering, high avidity antibodies whose association rate constant is less important than their dissociation rate constant.
Figures


References
-
- Dodoo D, Staalsoe T, Giha H, Kurtzhals JAL, Akanmori BD, Koram K, Dunyo S, Nkrumah FK, Hviid L, Theander TG. Antibodies to variant antigens on the surfaces of infected erythrocytes are associated with protection from malaria in Ghanaian children. Infect Immun. 2001;69:3713–3718. doi: 10.1128/IAI.69.6.3713-3718.2001. - DOI - PMC - PubMed
-
- Giha HA, Staalsoe T, Dodoo D, Roper C, Satti GM, Arnot DE, Hviid L, Theander TG. Antibodies to variable Plasmodium falciparum-infected erythrocyte surface antigens are associated with protection from novel malaria infections. Immunol Lett. 2000;71:117–126. doi: 10.1016/S0165-2478(99)00173-X. - DOI - PubMed
-
- Steckbeck JD, Orlov I, Chow A, Grieser H, Miller K, Bruno J, Robinson JE, Montelaro RC, Cole KS. Kinetic rates of antibody binding correlate with neutralization sensitivity of variant simian immunodeficiency virus strains. J Virol. 2005;79:12311–12320. doi: 10.1128/JVI.79.19.12311-12320.2005. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

