Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma
- PMID: 20403181
- PMCID: PMC2873438
- DOI: 10.1186/1476-4598-9-81
Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide with poor prognosis due to resistance to conventional chemotherapy and limited efficacy of radiotherapy. There is an urgent need to develop novel biomarkers for early diagnosis, as well as to identify new drug targets for therapeutic interventions.
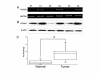
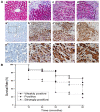
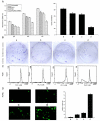
Patients and methods: 54 paired HCC samples and 21 normal liver tissues were obtained from West China Hospital of Sichuan University. Informed consent was obtained from all the patients or their relatives prior to analysis, and the project was approved by the Institutional Ethics Committee of Sichuan University. Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC)-based proteomics was employed to profile the differentially expressed proteins between a HepG2 human hepatoma cell line and an immortal hepatic cell line L02. Validation of PGAM1 expression was performed by semi-quantitative RT-PCR, immunoblot and immunohistochemistry using clinical samples. shRNA expressing plasmids specifically targeting PGAM1 were designed and constructed by GenePharma Corporation (Shanghai, China), and were utilized to silence expression of PGAM1 in vitro and in vivo. Cell proliferation was measured by a combination of colony formation assay and Ki67 staining. Apoptosis was examined by flow cytometry and TUNEL assay.
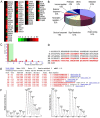
Results: A total of 63 dysregulated proteins were identified, including 51 up-regulated proteins, and 12 down-regulated proteins (over 2-fold, p < 0.01). Phosphoglycerate mutase 1 (PGAM1) was found markedly upregulated. Clinico-pathological analysis indicated that overexpression of PGAM1 was associated with 66.7% HCC, and strongly correlated with poor differentiation and decreased survival rates (p < 0.01). shRNAs-mediated repression of PGAM1 expression resulted in significant inhibition in liver cancer cell growth both in vitro and in vivo.
Conclusion: Our studies suggested that PGAM1 plays an important role in hepatocarcinogenesis, and should be a potential diagnostic biomarker, as well as an attractive therapeutic target for hepatocellular carcinoma.
Figures


Similar articles
-
Astragaloside IV inhibits cell viability and glycolysis of hepatocellular carcinoma by regulating KAT2A-mediated succinylation of PGAM1.BMC Cancer. 2024 Jun 4;24(1):682. doi: 10.1186/s12885-024-12438-9. BMC Cancer. 2024. PMID: 38835015 Free PMC article.
-
Phosphoglycerate mutase 1 knockdown inhibits prostate cancer cell growth, migration, and invasion.Asian J Androl. 2018 Mar-Apr;20(2):178-183. doi: 10.4103/aja.aja_57_17. Asian J Androl. 2018. PMID: 29271400 Free PMC article.
-
MicroRNA-27b exerts an oncogenic function by targeting Fbxw7 in human hepatocellular carcinoma.Tumour Biol. 2016 Nov;37(11):15325-15332. doi: 10.1007/s13277-016-5444-9. Epub 2016 Oct 4. Tumour Biol. 2016. PMID: 27704356
-
CD109 Mediates Cell Survival in Hepatocellular Carcinoma Cells.Dig Dis Sci. 2016 Aug;61(8):2303-2314. doi: 10.1007/s10620-016-4149-7. Epub 2016 Apr 13. Dig Dis Sci. 2016. Retraction in: Dig Dis Sci. 2019 Dec;64(12):3676. doi: 10.1007/s10620-019-05932-9. PMID: 27074923 Retracted.
-
Targeting PGAM1 in cancer: An emerging therapeutic opportunity.Eur J Med Chem. 2022 Dec 15;244:114798. doi: 10.1016/j.ejmech.2022.114798. Epub 2022 Oct 3. Eur J Med Chem. 2022. PMID: 36215859 Review.
Cited by
-
In-frame cDNA library combined with protein complementation assay identifies ARL11-binding partners.PLoS One. 2012;7(12):e52290. doi: 10.1371/journal.pone.0052290. Epub 2012 Dec 18. PLoS One. 2012. PMID: 23272234 Free PMC article.
-
Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question.Oncoscience. 2014 Dec 18;1(12):777-802. doi: 10.18632/oncoscience.109. eCollection 2014. Oncoscience. 2014. PMID: 25621294 Free PMC article.
-
Tumor microenvironment-activated cancer cell membrane-liposome hybrid nanoparticle-mediated synergistic metabolic therapy and chemotherapy for non-small cell lung cancer.J Nanobiotechnology. 2021 Oct 24;19(1):339. doi: 10.1186/s12951-021-01085-y. J Nanobiotechnology. 2021. PMID: 34689761 Free PMC article.
-
Single-cell landscape and spatial transcriptomic analysis reveals macrophage infiltration and glycolytic metabolism in kidney renal clear cell carcinoma.Aging (Albany NY). 2023 Oct 16;15(20):11298-11312. doi: 10.18632/aging.205128. Epub 2023 Oct 16. Aging (Albany NY). 2023. PMID: 37847178 Free PMC article.
-
Stress eating and tuning out: cancer cells re-wire metabolism to counter stress.Crit Rev Biochem Mol Biol. 2013 Nov-Dec;48(6):609-19. doi: 10.3109/10409238.2013.844093. Epub 2013 Oct 7. Crit Rev Biochem Mol Biol. 2013. PMID: 24099138 Free PMC article. Review.
References
-
- Chen Y, Lin MC, Yao H, Wang H, Zhang A, Yu J, Hui CK, Lau GK, He M, Sung J, Kung H. Lentivirus-Mediated RNA interference targeting enhancer of Zeste Homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of Stathmin. Hepatology. 2007;46:200–208. doi: 10.1002/hep.21668. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

