High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta
- PMID: 20403199
- PMCID: PMC2871261
- DOI: 10.1186/1471-2156-11-25
High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta
Abstract
Background: Imprinted genes show expression from one parental allele only and are important for development and behaviour. This extreme mode of allelic imbalance has been described for approximately 56 human genes. Imprinting status is often disrupted in cancer and dysmorphic syndromes. More subtle variation of gene expression, that is not parent-of-origin specific, termed 'allele-specific gene expression' (ASE) is more common and may give rise to milder phenotypic differences. Using two allele-specific high-throughput technologies alongside bioinformatics predictions, normal term human placenta was screened to find new imprinted genes and to ascertain the extent of ASE in this tissue.
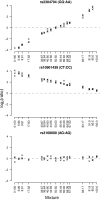
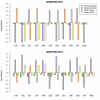
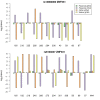
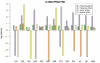
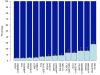
Results: Twenty-three family trios of placental cDNA, placental genomic DNA (gDNA) and gDNA from both parents were tested for 130 candidate genes with the Sequenom MassArray system. Six genes were found differentially expressed but none imprinted. The Illumina ASE BeadArray platform was then used to test 1536 SNPs in 932 genes. The array was enriched for the human orthologues of 124 mouse candidate genes from bioinformatics predictions and 10 human candidate imprinted genes from EST database mining. After quality control pruning, a total of 261 informative SNPs (214 genes) remained for analysis. Imprinting with maternal expression was demonstrated for the lymphocyte imprinted gene ZNF331 in human placenta. Two potential differentially methylated regions (DMRs) were found in the vicinity of ZNF331. None of the bioinformatically predicted candidates tested showed imprinting except for a skewed allelic expression in a parent-specific manner observed for PHACTR2, a neighbour of the imprinted PLAGL1 gene. ASE was detected for two or more individuals in 39 candidate genes (18%).
Conclusions: Both Sequenom and Illumina assays were sensitive enough to study imprinting and strong allelic bias. Previous bioinformatics approaches were not predictive of new imprinted genes in the human term placenta. ZNF331 is imprinted in human term placenta and might be a new ubiquitously imprinted gene, part of a primate-specific locus. Demonstration of partial imprinting of PHACTR2 calls for re-evaluation of the allelic pattern of expression for the PHACTR2-PLAGL1 locus. ASE was common in human term placenta.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

