Influenza A viral loads in respiratory samples collected from patients infected with pandemic H1N1, seasonal H1N1 and H3N2 viruses
- PMID: 20403211
- PMCID: PMC2874536
- DOI: 10.1186/1743-422X-7-75
Influenza A viral loads in respiratory samples collected from patients infected with pandemic H1N1, seasonal H1N1 and H3N2 viruses
Abstract
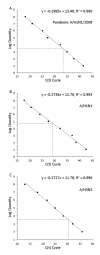
Background: Nasopharyngeal aspirate (NPA), nasal swab (NS), and throat swab (TS) are common specimens used for diagnosis of respiratory virus infections based on the detection of viral genomes, viral antigens and viral isolation. However, there is no documented data regarding the type of specimen that yields the best result of viral detection. In this study, quantitative real time RT-PCR specific for M gene was used to determine influenza A viral loads present in NS, NPA and TS samples collected from patients infected with the 2009 pandemic H1N1, seasonal H1N1 and H3N2 viruses. Various copy numbers of RNA transcripts derived from recombinant plasmids containing complete M gene insert of each virus strain were assayed by RT-PCR. A standard curve for viral RNA quantification was constructed by plotting each Ct value against the log quantity of each standard RNA copy number.
Results: Copy numbers of M gene were obtained through the extrapolation of Ct values of the test samples against the corresponding standard curve. Among a total of 29 patients with severe influenza enrolled in this study (12 cases of the 2009 pandemic influenza, 5 cases of seasonal H1N1 and 12 cases of seasonal H3N2 virus), NPA was found to contain significantly highest amount of viral loads and followed in order by NS and TS specimen. Viral loads among patients infected with those viruses were comparable regarding type of specimen analyzed.
Conclusion: Based on M gene copy numbers, we conclude that NPA is the best specimen for detection of influenza A viruses, and followed in order by NS and TS.
Figures

References
-
- WHO information for laboratory diagnosis of pandemic (H1N1) 2009 virus in humans-update. http://www.who.int/csr/resources/publications/swineflu/en/
-
- CDC protocol of realtime RTPCR for influenza A (H1N1) http://www.who.int/csr/resources/publications/swineflu/en/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

