Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator
- PMID: 20403323
- PMCID: PMC4549071
- DOI: 10.1016/j.cell.2010.03.021
Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator
Abstract
One oscillation of Cyclin-dependent kinase (Cdk) activity, largely driven by periodic synthesis and destruction of cyclins, is tightly coupled to a single complete eukaryotic cell division cycle. Tight linkage of different steps in diverse cell-cycle processes to Cdk activity has been proposed to explain this coupling. Here, we demonstrate an intrinsically oscillatory module controlling nucleolar release and resequestration of the Cdc14 phosphatase, which is essential for mitotic exit in budding yeast. We find that this Cdc14 release oscillator functions at constant and physiological cyclin-Cdk levels, and is therefore independent of the Cdk oscillator. However, the frequency of the release oscillator is regulated by cyclin-Cdk activity. This observation together with its mechanism suggests that the intrinsically autonomous Cdc14 release cycles are locked at once-per-cell-cycle through entrainment by the Cdk oscillator in wild-type cells. This concept may have broad implications for the structure and evolution of eukaryotic cell-cycle control.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

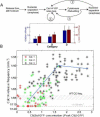
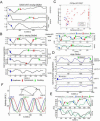
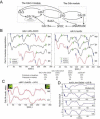
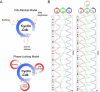
Comment in
-
The hidden rhythms of the dividing cell.Cell. 2010 Apr 16;141(2):224-6. doi: 10.1016/j.cell.2010.03.042. Cell. 2010. PMID: 20403319
Similar articles
-
Oscillations in Cdc14 release and sequestration reveal a circuit underlying mitotic exit.J Cell Biol. 2010 Jul 26;190(2):209-22. doi: 10.1083/jcb.201002026. J Cell Biol. 2010. PMID: 20660629 Free PMC article.
-
Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus.Science. 2004 Jul 23;305(5683):516-9. doi: 10.1126/science.1099402. Science. 2004. PMID: 15273393
-
The molecular function of the yeast polo-like kinase Cdc5 in Cdc14 release during early anaphase.Mol Biol Cell. 2009 Aug;20(16):3671-9. doi: 10.1091/mbc.e08-10-1049. Epub 2009 Jul 1. Mol Biol Cell. 2009. PMID: 19570916 Free PMC article.
-
The Multiple Roles of the Cdc14 Phosphatase in Cell Cycle Control.Int J Mol Sci. 2020 Jan 21;21(3):709. doi: 10.3390/ijms21030709. Int J Mol Sci. 2020. PMID: 31973188 Free PMC article. Review.
-
Decoding the Nucleolar Role in Meiotic Recombination and Cell Cycle Control: Insights into Cdc14 Function.Int J Mol Sci. 2024 Nov 29;25(23):12861. doi: 10.3390/ijms252312861. Int J Mol Sci. 2024. PMID: 39684572 Free PMC article. Review.
Cited by
-
Degradation of the Mitotic Cyclin Clb3 Is not Required for Mitotic Exit but Is Necessary for G1 Cyclin Control of the Succeeding Cell Cycle.Genetics. 2016 Dec;204(4):1479-1494. doi: 10.1534/genetics.116.194837. Epub 2016 Oct 28. Genetics. 2016. PMID: 27794027 Free PMC article.
-
Frequency control of cell cycle oscillators.Curr Opin Genet Dev. 2010 Dec;20(6):605-12. doi: 10.1016/j.gde.2010.08.006. Epub 2010 Sep 28. Curr Opin Genet Dev. 2010. PMID: 20851595 Free PMC article. Review.
-
Principles, mechanisms and functions of entrainment in biological oscillators.Interface Focus. 2022 Apr 15;12(3):20210088. doi: 10.1098/rsfs.2021.0088. eCollection 2022 Jun 6. Interface Focus. 2022. PMID: 35450280 Free PMC article. Review.
-
A transcriptome-wide analysis deciphers distinct roles of G1 cyclins in temporal organization of the yeast cell cycle.Sci Rep. 2019 Mar 4;9(1):3343. doi: 10.1038/s41598-019-39850-7. Sci Rep. 2019. PMID: 30833602 Free PMC article.
-
Phase resetting reveals network dynamics underlying a bacterial cell cycle.PLoS Comput Biol. 2012;8(11):e1002778. doi: 10.1371/journal.pcbi.1002778. Epub 2012 Nov 29. PLoS Comput Biol. 2012. PMID: 23209388 Free PMC article.
References
-
- Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science (New York, NY. 2004;305:516–519. - PubMed
-
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. - PubMed
-
- Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Molecular cell. 2006;21:3–14. - PubMed
-
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annual review of biochemistry. 2006;75:655–680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

