The tri-nucleotide spacer sequence between estrogen response element half-sites is conserved and modulates ERalpha-mediated transcriptional responses
- PMID: 20403436
- PMCID: PMC2891080
- DOI: 10.1016/j.jsbmb.2010.04.009
The tri-nucleotide spacer sequence between estrogen response element half-sites is conserved and modulates ERalpha-mediated transcriptional responses
Abstract
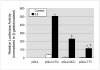
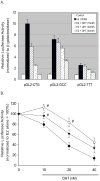
The estrogen response element (ERE) consensus sequence is AGGTCAnnnTGACCT, where nnn is known as the tri-nucleotide spacer sequence. Studying 1017 high-confidence ERalpha-bound loci, we found that genomic EREs are enriched for spacers composed of C(A/T)G, suggesting that the spacer may influence receptor binding and transcriptional responses. We designed consensus EREs containing variable spacer sequences and compared ERalpha binding in gel shift assays and enhancer function in reporter assays. We found that ERalpha-ERE binding affinity is modulated by the tri-nucleotide spacer sequence and is favored by spacer sequences of CTG>GCC>TTT. Similarly, luciferase reporter assays indicated that the estrogen-stimulated transcriptional response is modulated by the spacer and parallels the gel shift data: CTG>GCC>TTT. Reporter assays demonstrated that the spacer sequence also modulates the sensitivity of EREs to repression engendered by the receptor antagonist hydroxytamoxifen. These experiments indicate that the sequence of the tri-nucleotide spacer is non-random at receptor-bound genomic loci, influences ERalpha-DNA-binding affinity, and modulates transactivation potential of the receptor-ligand-DNA complex. This work has implications for understanding which genomic EREs are targeted by ERalpha, should improve computational prediction of functional EREs within genomic sequences, and describes novel sequence determinants of the estrogen response.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

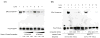
References
-
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354(3):270–282. - PubMed
-
- Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. - PubMed
-
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842. - PubMed
-
- Cerillo G, Rees A, Manchanda N, Reilly C, Brogan I, White A, Needham M. The oestrogen receptor regulates NFkappaB and AP-1 activity in a cell-specific manner. J Steroid Biochem Mol Biol. 1998;67(2):79–88. - PubMed
-
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277(5331):1508–1510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

