Two-step imprinted X inactivation: repeat versus genic silencing in the mouse
- PMID: 20404085
- PMCID: PMC2897575
- DOI: 10.1128/MCB.00227-10
Two-step imprinted X inactivation: repeat versus genic silencing in the mouse
Abstract
Mammals compensate for unequal X-linked gene dosages between the sexes by inactivating one X chromosome in the female. In marsupials and in the early mouse embryo, X chromosome inactivation (XCI) is imprinted to occur selectively on the paternal X chromosome (X(P)). The mechanisms and events underlying X(P) imprinting remain unclear. Here, we find that the imprinted X(P) can be functionally divided into two domains, one comprising traditional coding genes (genic) and the other comprising intergenic repetitive elements. X(P) repetitive element silencing occurs by the two-cell stage, does not require Xist, and occurs several divisions prior to genic silencing. In contrast, genic silencing initiates at the morula-to-blastocyst stage and absolutely requires Xist. Genes translocate into the presilenced repeat region as they are inactivated, whereas active genes remain outside. Thus, during the gamete-embryo transition, imprinted XCI occurs in two steps, with repeat silencing preceding genic inactivation. Nucleolar association may underlie the epigenetic asymmetry of X(P) and X(M). We hypothesize that transgenerational information (the imprint) is carried by repeats from the paternal germ line or that, alternatively, repetitive elements are silenced at the two-cell stage in a parent-of-origin-specific manner. Our model incorporates aspects of the so-called classical, de novo, and preinactivation hypotheses and suggests that Xist RNA functions relatively late during preimplantation mouse development.
Figures


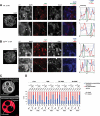
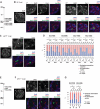

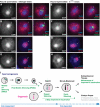
References
-
- Adler, D., J. West, and V. Chapman. 1977. Expression of alpha-galactosidase in preimplantation mouse embryos. Nature 267:838-839. - PubMed
-
- Avner, P., and E. Heard. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2:59-67. - PubMed
-
- Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 81:493-525. - PubMed
-
- Beatty, B., S. Mai, and J. Squire. 2002. FISH. A practical approach. Oxford University Press, Oxford, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
