Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes
- PMID: 20404116
- PMCID: PMC2897269
- DOI: 10.1128/AAC.01834-09
Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes
Abstract
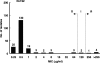
We evaluated the in vitro activity of fosfomycin against a total of 192 CTX-M beta-lactamase-producing Escherichia coli strains isolated in 70 Japanese clinical settings. Most of the isolates (96.4%) were found to be susceptible to fosfomycin. On the other hand, some of the resistant isolates were confirmed to harbor the novel transferable fosfomycin resistance determinants named FosA3 and FosC2, which efficaciously inactivate fosfomycin through glutathione S-transferase activity.
Figures
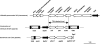

References
-
- Beharry, Z., and T. Palzkill. 2005. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J. Biol. Chem. 280:17786-17791. - PubMed
-
- Bernat, B. A., L. T. Laughlin, and R. N. Armstrong. 1999. Elucidation of a monovalent cation dependence and characterization of the divalent cation binding site of the fosfomycin resistance protein (FosA). Biochemistry 38:7462-7469. - PubMed
-
- Bernat, B. A., L. T. Laughlin, and R. N. Armstrong. 1997. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry 36:3050-3055. - PubMed
-
- Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 8th ed. Document M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
-
- Endimiani, A., G. Patel, K. M. Hujer, M. Swaminathan, F. Perez, L. B. Rice, M. R. Jacobs, and R. A. Bonomo. 2009. In vitro activity of fosfomycin against blaKPC-containing Klebsiella pneumoniae isolates, including those nonsusceptible to tigecycline and/or colistin. Antimicrob. Agents Chemother. 54:526-529. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

