HIV-1 protease codon 36 polymorphisms and differential development of resistance to nelfinavir, lopinavir, and atazanavir in different HIV-1 subtypes
- PMID: 20404123
- PMCID: PMC2897293
- DOI: 10.1128/AAC.01828-09
HIV-1 protease codon 36 polymorphisms and differential development of resistance to nelfinavir, lopinavir, and atazanavir in different HIV-1 subtypes
Abstract
The amino acid at position 36 of the HIV-1 protease differs among various viral subtypes, in that methionine is usually found in subtype B viruses but isoleucine is common in other subtypes. This polymorphism is associated with higher rates of treatment failure involving protease inhibitors (PIs) in non-subtype B-infected patients. To investigate this, we generated genetically homogeneous wild-type viruses from subtype B, subtype C, and CRF02_AG full-length molecular clones and showed that subtype C and CRF02_AG I36 viruses exhibited higher levels of resistance to various PIs than their respective M36 counterparts, while the opposite was observed for subtype B viruses. Selections for resistance with each variant were performed with nelfinavir (NFV), lopinavir (LPV), and atazanavir (ATV). Sequence analysis of the protease gene at week 35 revealed that the major NFV resistance mutation D30N emerged in NFV-selected subtype B viruses and in I36 subtype C viruses, despite polymorphic variation. A unique mutational pattern developed in subtype C M36 viruses selected with NFV or ATV. The presence of I47A in LPV-selected I36 CRF02_AG virus conferred higher-level resistance than L76V in LPV-selected M36 CRF02_AG virus. Phenotypic analysis revealed a >1,000-fold increase in NFV resistance in I36 subtype C NFV-selected virus with no apparent impact on viral replication capacity. Thus, the position 36 polymorphism in the HIV-1 protease appears to have a differential effect on both drug susceptibility and the viral replication capacity, depending on both the viral subtype and the drug being evaluated.
Figures

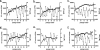
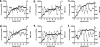
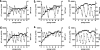
Similar articles
-
The role of polymorphisms at position 89 in the HIV-1 protease gene in the development of drug resistance to HIV-1 protease inhibitors.J Antimicrob Chemother. 2012 Apr;67(4):988-94. doi: 10.1093/jac/dkr582. Epub 2012 Feb 7. J Antimicrob Chemother. 2012. PMID: 22315096
-
Mutational patterns and correlated amino acid substitutions in the HIV-1 protease after virological failure to nelfinavir- and lopinavir/ritonavir-based treatments.J Med Virol. 2007 Nov;79(11):1617-28. doi: 10.1002/jmv.20986. J Med Virol. 2007. PMID: 17854027
-
Impact of frequent natural polymorphisms at the protease gene on the in vitro susceptibility to protease inhibitors in HIV-1 non-B subtypes.J Clin Virol. 2004 Nov;31(3):215-20. doi: 10.1016/j.jcv.2004.03.015. J Clin Virol. 2004. PMID: 15465415
-
Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution.AIDS Rev. 2003 Jan-Mar;5(1):25-35. AIDS Rev. 2003. PMID: 12875105 Review.
-
Antitumorigenic action of nelfinavir: Effects on multiple myeloma and hematologic malignancies (Review).Oncol Rep. 2020 Jun;43(6):1729-1736. doi: 10.3892/or.2020.7562. Epub 2020 Mar 26. Oncol Rep. 2020. PMID: 32236596 Review.
Cited by
-
Comparable long-term efficacy of Lopinavir/Ritonavir and similar drug-resistance profiles in different HIV-1 subtypes.PLoS One. 2014 Jan 27;9(1):e86239. doi: 10.1371/journal.pone.0086239. eCollection 2014. PLoS One. 2014. PMID: 24475093 Free PMC article.
-
Role of HIV Subtype Diversity in the Development of Resistance to Antiviral Drugs.Viruses. 2010 Nov;2(11):2493-508. doi: 10.3390/v2112493. Epub 2010 Nov 11. Viruses. 2010. PMID: 21994627 Free PMC article.
-
Key Factors Influencing the Emergence of Human Immunodeficiency Virus Drug Resistance in Low- and Middle-Income Countries.J Infect Dis. 2017 Dec 1;216(suppl_9):S851-S856. doi: 10.1093/infdis/jix409. J Infect Dis. 2017. PMID: 29207000 Free PMC article. Review.
-
Comparative biochemical analysis of recombinant reverse transcriptase enzymes of HIV-1 subtype B and subtype C.Retrovirology. 2010 Oct 7;7:80. doi: 10.1186/1742-4690-7-80. Retrovirology. 2010. PMID: 20929562 Free PMC article.
-
Characterization of the Drug Resistance Profiles of Patients Infected with CRF07_BC Using Phenotypic Assay and Ultra-Deep Pyrosequencing.PLoS One. 2017 Jan 20;12(1):e0170420. doi: 10.1371/journal.pone.0170420. eCollection 2017. PLoS One. 2017. PMID: 28107423 Free PMC article.
References
-
- Bessong, P. O. 2008. Polymorphisms in HIV-1 subtype C proteases and the potential impact on protease inhibitors. Trop. Med. Int. Health 13:144-151. - PubMed
-
- Brenner, B., D. Turner, M. Oliveira, D. Moisi, M. Detorio, M. Carobene, R. G. Marlink, J. Schapiro, M. Roger, and M. A. Wainberg. 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17:F1-F5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

