Downstream targets of HOXB4 in a cell line model of primitive hematopoietic progenitor cells
- PMID: 20404135
- PMCID: PMC2918329
- DOI: 10.1182/blood-2009-11-253872
Downstream targets of HOXB4 in a cell line model of primitive hematopoietic progenitor cells
Abstract
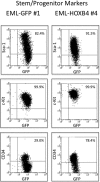
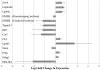
Enforced expression of the homeobox transcription factor HOXB4 has been shown to enhance hematopoietic stem cell self-renewal and expansion ex vivo and in vivo. To investigate the downstream targets of HOXB4 in hematopoietic progenitor cells, HOXB4 was constitutively overexpressed in the primitive hematopoietic progenitor cell line EML. Two genome-wide analytical techniques were used: RNA expression profiling using microarrays and chromatin immunoprecipitation (ChIP)-chip. RNA expression profiling revealed that 465 gene transcripts were differentially expressed in KLS (c-Kit(+), Lin(-), Sca-1(+))-EML cells that overexpressed HOXB4 (KLS-EML-HOXB4) compared with control KLS-EML cells that were transduced with vector alone. In particular, erythroid-specific gene transcripts were observed to be highly down-regulated in KLS-EML-HOXB4 cells. ChIP-chip analysis revealed that the promoter region for 1910 genes, such as CD34, Sox4, and B220, were occupied by HOXB4 in KLS-EML-HOXB4 cells. Side-by-side comparison of the ChIP-chip and RNA expression profiling datasets provided correlative information and identified Gp49a and Laptm4b as candidate "stemness-related" genes. Both genes were highly ranked in both dataset lists and have been previously shown to be preferentially expressed in hematopoietic stem cells and down-regulated in mature hematopoietic cells, thus making them attractive candidates for future functional studies in hematopoietic cells.
Figures




Similar articles
-
Hemgn is a direct transcriptional target of HOXB4 and induces expansion of murine myeloid progenitor cells.Blood. 2010 Aug 5;116(5):711-9. doi: 10.1182/blood-2009-07-235341. Epub 2010 Apr 14. Blood. 2010. PMID: 20393131 Free PMC article.
-
Enhanced in vivo regenerative potential of HOXB4-transduced hematopoietic stem cells with regulation of their pool size.Blood. 1999 Oct 15;94(8):2605-12. Blood. 1999. PMID: 10515864
-
NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4.Blood. 2003 Oct 1;102(7):2420-7. doi: 10.1182/blood-2003-01-0251. Epub 2003 Jun 5. Blood. 2003. PMID: 12791656
-
[Ex vivo expansion of human hematopoietic stem cells by passive transduction of the HOXB4 homeoprotein].J Soc Biol. 2006;200(3):235-41. doi: 10.1051/jbio:2006027. J Soc Biol. 2006. PMID: 17417138 Review. French.
-
[Regulatory role of HOXB4 in self-renewal of hematopoietic stem cells - review].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2007 Jun;15(3):647-51. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2007. PMID: 17605886 Review. Chinese.
Cited by
-
Dynamic HoxB4-regulatory network during embryonic stem cell differentiation to hematopoietic cells.Blood. 2012 May 10;119(19):e139-47. doi: 10.1182/blood-2011-12-396754. Epub 2012 Mar 21. Blood. 2012. PMID: 22438249 Free PMC article.
-
Selection of Leptin Surrogates by a General Phenotypic Screening Method for Receptor Agonists.Biomolecules. 2024 Apr 9;14(4):457. doi: 10.3390/biom14040457. Biomolecules. 2024. PMID: 38672473 Free PMC article.
-
Densely interconnected transcriptional circuits control cell states in human hematopoiesis.Cell. 2011 Jan 21;144(2):296-309. doi: 10.1016/j.cell.2011.01.004. Cell. 2011. PMID: 21241896 Free PMC article.
-
Correlation of LAPTM4B polymorphisms with gallbladder carcinoma susceptibility in Chinese patients.Med Oncol. 2012 Dec;29(4):2809-13. doi: 10.1007/s12032-012-0173-4. Med Oncol. 2012. PMID: 22302286
-
Future perspectives: therapeutic targeting of notch signalling may become a strategy in patients receiving stem cell transplantation for hematologic malignancies.Bone Marrow Res. 2011;2011:570796. doi: 10.1155/2011/570796. Epub 2010 Oct 4. Bone Marrow Res. 2011. PMID: 22046566 Free PMC article.
References
-
- Chao NJ, Emerson SG, Weinberg KI. Stem cell transplantation (cord blood transplants). Hematology Am Soc Hematol Educ Program. 2004:354–371. - PubMed
-
- Sorrentino BP. Clinical strategies for expansion of haematopoietic stem cells. Nat Rev Immunol. 2004;4(11):878–888. - PubMed
-
- Bhardwaj G, Murdoch B, Wu D, et al. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol. 2001;2(2):172–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32-HL07262/HL/NHLBI NIH HHS/United States
- P30 DK072442/DK/NIDDK NIH HHS/United States
- K08 DK064809/DK/NIDDK NIH HHS/United States
- T32 HD007149/HD/NICHD NIH HHS/United States
- P30-DK72442/DK/NIDDK NIH HHS/United States
- P30-CA016359/CA/NCI NIH HHS/United States
- T32-HD07149/HD/NICHD NIH HHS/United States
- P01 HL063357/HL/NHLBI NIH HHS/United States
- T32 HL007262/HL/NHLBI NIH HHS/United States
- P30 CA016359/CA/NCI NIH HHS/United States
- P01-HL63357/HL/NHLBI NIH HHS/United States
- K08-DK064809/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

