Genetic suppression of the circadian Clock mutation by the melatonin biosynthesis pathway
- PMID: 20404168
- PMCID: PMC2889547
- DOI: 10.1073/pnas.1004368107
Genetic suppression of the circadian Clock mutation by the melatonin biosynthesis pathway
Abstract
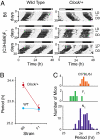
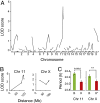
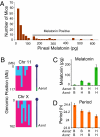
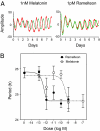
Most laboratory mouse strains including C57BL/6J do not produce detectable levels of pineal melatonin owing to deficits in enzymatic activity of arylalkylamine N-acetyltransferase (AANAT) and N-acetylserotonin O-methyl transferase (ASMT), two enzymes necessary for melatonin biosynthesis. Here we report that alleles segregating at these two loci in C3H/HeJ mice, an inbred strain producing melatonin, suppress the circadian period-lengthening effect of the Clock mutation. Through a functional mapping approach, we localize mouse Asmt to chromosome X and show that it, and the Aanat locus on chromosome 11, are significantly associated with pineal melatonin levels. Treatment of suprachiasmatic nucleus (SCN) explant cultures from Period2(Luciferase) (Per2(Luc)) Clock/+ reporter mice with melatonin, or the melatonin agonist, ramelteon, phenocopies the genetic suppression of the Clock mutant phenotype observed in living animals. These results demonstrate that melatonin suppresses the Clock/+ mutant phenotype and interacts with Clock to affect the mammalian circadian system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A congenic line of the C57BL/6J mouse strain that is proficient in melatonin synthesis.J Pineal Res. 2018 Oct;65(3):e12509. doi: 10.1111/jpi.12509. Epub 2018 Jun 6. J Pineal Res. 2018. PMID: 29768727 Free PMC article.
-
Season-dependent postembryonic maturation of the diurnal rhythm of melatonin biosynthesis in the chicken pineal gland.Chronobiol Int. 2012 Nov;29(9):1227-38. doi: 10.3109/07420528.2012.719964. Epub 2012 Sep 24. Chronobiol Int. 2012. PMID: 23003334
-
The Absence of Pineal Melatonin Abolishes the Daily Rhythm of Tph1 (Tryptophan Hydroxylase 1), Asmt (Acetylserotonin O-Methyltransferase), and Aanat (Aralkylamine N-Acetyltransferase) mRNA Expressions in Rat Testes.Mol Neurobiol. 2019 Nov;56(11):7800-7809. doi: 10.1007/s12035-019-1626-y. Epub 2019 May 23. Mol Neurobiol. 2019. PMID: 31124080
-
Melatonin synthesis pathway: circadian regulation of the genes encoding the key enzymes in the chicken pineal gland and retina.Reprod Nutr Dev. 1999 May-Jun;39(3):325-34. doi: 10.1051/rnd:19990305. Reprod Nutr Dev. 1999. PMID: 10420435 Review.
-
Melatonin synthesis and clock gene regulation in the pineal organ of teleost fish compared to mammals: Similarities and differences.Gen Comp Endocrinol. 2019 Aug 1;279:27-34. doi: 10.1016/j.ygcen.2018.07.010. Epub 2018 Jul 17. Gen Comp Endocrinol. 2019. PMID: 30026020 Review.
Cited by
-
Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock Δ19/+ mice contributes to improved glucose homeostasis.Sci Rep. 2015 Jul 31;5:12801. doi: 10.1038/srep12801. Sci Rep. 2015. PMID: 26228022 Free PMC article.
-
Circadian regulation of pineal gland rhythmicity.Mol Cell Endocrinol. 2012 Feb 5;349(1):13-9. doi: 10.1016/j.mce.2011.07.009. Epub 2011 Jul 18. Mol Cell Endocrinol. 2012. PMID: 21782887 Free PMC article. Review.
-
Influence of photoperiod on hormones, behavior, and immune function.Front Neuroendocrinol. 2011 Aug;32(3):303-19. doi: 10.1016/j.yfrne.2010.12.003. Epub 2010 Dec 13. Front Neuroendocrinol. 2011. PMID: 21156187 Free PMC article. Review.
-
Melatonin Does Not Affect the Stress-Induced Phase Shifts of Peripheral Clocks in Male Mice.Endocrinology. 2023 Nov 20;165(1):bqad183. doi: 10.1210/endocr/bqad183. Endocrinology. 2023. PMID: 38128120 Free PMC article.
-
CLOCKΔ19 mutation modifies the manner of synchrony among oscillation neurons in the suprachiasmatic nucleus.Sci Rep. 2018 Jan 16;8(1):854. doi: 10.1038/s41598-018-19224-1. Sci Rep. 2018. PMID: 29339832 Free PMC article.
References
-
- Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. - PubMed
-
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

