A forward genetic screen in Drosophila melanogaster to identify mutations affecting INAD localization in photoreceptor cells
- PMID: 20404479
- PMCID: PMC7505561
- DOI: 10.4161/fly.4.2.11861
A forward genetic screen in Drosophila melanogaster to identify mutations affecting INAD localization in photoreceptor cells
Abstract
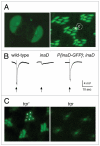
In Drosophila photoreceptors, the multivalent PDZ protein INAD interacts with multiple signaling components and localizes complexes to the rhabdomere, a subcellular compartment specialized for phototransduction. Since this localization is critical for signaling, we conducted a genetic screen of the third chromosome for mutations that result in mislocalization of an INAD-GFP fusion protein. We identified seven mutant lines that fall into two complementation groups, idl (INAD localization)-A and idl-B. We show that idl-A mutants fail to complement with chaoptic (chp) mutants. Since chaoptin is a structural component of the rhabdomere, mislocalization of INAD may be a secondary effect of the retinal degeneration in chp and idl-A mutants. Genetic complementation and DNA sequencing reveal that the two idl-B mutants represent new alleles of trp, a gene encoding the major light-activated channel. The molecular change in each allele affects a highly conserved residue in either an ankyrin domain on the N-terminus or in the S6 transmembrane domain of TRP. These changes lead to the loss of TRP protein. TRP has previously been shown to anchor INAD in the rhabdomeres, therefore the independent identification of two trp alleles validates our screen for INAD-GFP localization. One possibility is that a limited number of proteins are required for localizing INAD-signaling complexes. A similar screen of the X and second chromosomes may be required to find the remaining players involved.
Figures






Similar articles
-
Two distantly positioned PDZ domains mediate multivalent INAD-phospholipase C interactions essential for G protein-coupled signaling.EMBO J. 1998 Apr 15;17(8):2285-97. doi: 10.1093/emboj/17.8.2285. EMBO J. 1998. PMID: 9545241 Free PMC article.
-
Interaction of eye protein kinase C and INAD in Drosophila. Localization of binding domains and electrophysiological characterization of a loss of association in transgenic flies.J Biol Chem. 1998 Jul 10;273(28):17713-9. doi: 10.1074/jbc.273.28.17713. J Biol Chem. 1998. PMID: 9651370
-
Independent anchoring and assembly mechanisms of INAD signaling complexes in Drosophila photoreceptors.J Neurosci. 2001 Jan 1;21(1):150-8. doi: 10.1523/JNEUROSCI.21-01-00150.2001. J Neurosci. 2001. PMID: 11150331 Free PMC article.
-
Molecular genetics of Drosophila TRP channels.Novartis Found Symp. 2004;258:3-12; discussion 12-7, 98-102, 263-6. Novartis Found Symp. 2004. PMID: 15104173 Review.
-
Phototransduction and retinal degeneration in Drosophila.Pflugers Arch. 2007 Aug;454(5):821-47. doi: 10.1007/s00424-007-0251-1. Epub 2007 May 9. Pflugers Arch. 2007. PMID: 17487503 Review.
Cited by
-
chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells.Biol Open. 2014 Apr 4;3(5):332-41. doi: 10.1242/bio.20147310. Biol Open. 2014. PMID: 24705015 Free PMC article.
References
-
- Tsunoda S, Zuker CS. The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling. Cell Calcium. 1999;26:165–71. - PubMed
-
- Huber A. Scaffolding proteins organize multimolecular protein complexes for sensory signal transduction. The European journal of neuroscience. 2001;14:769–76. - PubMed
-
- Ranganathan R, Ross EM. PDZ domain proteins: scaffolds for signaling complexes. Curr Biol. 1997;7:770–3. - PubMed
-
- Tsunoda S, Sierralta J, Zuker CS. Specificity in signaling pathways: assembly into multimolecular signaling complexes. Curr Opin Genet Dev. 1998;8:419–22. - PubMed
-
- Hardie RC, Raghu P. Visual transduction in Drosophila. Nature. 2001;413:186–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
