Identification of SOX9 interaction sites in the genome of chondrocytes
- PMID: 20404928
- PMCID: PMC2852419
- DOI: 10.1371/journal.pone.0010113
Identification of SOX9 interaction sites in the genome of chondrocytes
Abstract
Background: Our previous work has provided strong evidence that the transcription factor SOX9 is completely needed for chondrogenic differentiation and cartilage formation acting as a "master switch" in this differentiation. Heterozygous mutations in SOX9 cause campomelic dysplasia, a severe skeletal dysmorphology syndrome in humans characterized by a generalized hypoplasia of endochondral bones. To obtain insights into the logic used by SOX9 to control a network of target genes in chondrocytes, we performed a ChIP-on-chip experiment using SOX9 antibodies.
Methodology/principal findings: The ChIP DNA was hybridized to a microarray, which covered 80 genes, many of which are involved in chondrocyte differentiation. Hybridization peaks were detected in a series of cartilage extracellular matrix (ECM) genes including Col2a1, Col11a2, Aggrecan and Cdrap as well as in genes for specific transcription factors and signaling molecules. Our results also showed SOX9 interaction sites in genes that code for proteins that enhance the transcriptional activity of SOX9. Interestingly, a strong SOX9 signal was also observed in genes such as Col1a1 and Osx, whose expression is strongly down regulated in chondrocytes but is high in osteoblasts. In the Col2a1 gene, in addition to an interaction site on a previously identified enhancer in intron 1, another strong interaction site was seen in intron 6. This site is free of nucleosomes specifically in chondrocytes suggesting an important role of this site on Col2a1 transcription regulation by SOX9.
Conclusions/significance: Our results provide a broad understanding of the strategies used by a "master" transcription factor of differentiation in control of the genetic program of chondrocytes.
Conflict of interest statement
Figures

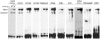



Similar articles
-
L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway.Osteoarthritis Cartilage. 2001;9 Suppl A:S69-75. doi: 10.1053/joca.2001.0447. Osteoarthritis Cartilage. 2001. PMID: 11680692
-
SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression.PLoS Genet. 2011 Nov;7(11):e1002356. doi: 10.1371/journal.pgen.1002356. Epub 2011 Nov 3. PLoS Genet. 2011. PMID: 22072985 Free PMC article.
-
SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene.Mol Cell Biol. 1997 Apr;17(4):2336-46. doi: 10.1128/MCB.17.4.2336. Mol Cell Biol. 1997. PMID: 9121483 Free PMC article.
-
Transcriptional mechanisms of chondrocyte differentiation.Matrix Biol. 2000 Sep;19(5):389-94. doi: 10.1016/s0945-053x(00)00094-9. Matrix Biol. 2000. PMID: 10980415 Review.
-
SOX9 and the many facets of its regulation in the chondrocyte lineage.Connect Tissue Res. 2017 Jan;58(1):2-14. doi: 10.1080/03008207.2016.1183667. Epub 2016 Apr 29. Connect Tissue Res. 2017. PMID: 27128146 Free PMC article. Review.
Cited by
-
The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis.Nucleic Acids Res. 2015 Sep 30;43(17):8183-203. doi: 10.1093/nar/gkv688. Epub 2015 Jul 6. Nucleic Acids Res. 2015. PMID: 26150426 Free PMC article.
-
Smpd3 Expression in both Chondrocytes and Osteoblasts Is Required for Normal Endochondral Bone Development.Mol Cell Biol. 2016 Aug 12;36(17):2282-99. doi: 10.1128/MCB.01077-15. Print 2016 Sep 1. Mol Cell Biol. 2016. PMID: 27325675 Free PMC article.
-
Disruption of SATB2 or its long-range cis-regulation by SOX9 causes a syndromic form of Pierre Robin sequence.Hum Mol Genet. 2014 May 15;23(10):2569-79. doi: 10.1093/hmg/ddt647. Epub 2013 Dec 20. Hum Mol Genet. 2014. PMID: 24363063 Free PMC article.
-
SOX9: a stem cell transcriptional regulator of secreted niche signaling factors.Genes Dev. 2014 Feb 15;28(4):328-41. doi: 10.1101/gad.233247.113. Genes Dev. 2014. PMID: 24532713 Free PMC article.
-
Disorders of sex development: new genes, new concepts.Nat Rev Endocrinol. 2013 Feb;9(2):79-91. doi: 10.1038/nrendo.2012.235. Epub 2012 Dec 18. Nat Rev Endocrinol. 2013. PMID: 23296159 Review.
References
-
- Chaboissier MC, Kobayashi A, Vidal Vl, Lützkendorf S, van de Kant HJ, et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. - PubMed
-
- Vidal VP, Chaboissier MC, Lützkendorf S, Cotsarelis G, Mill P, et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol. 2005;15:1340–1351. - PubMed
-
- Mori-Akiyama Y, van den Born M, van Es JH, Hamilton SR, Adams HP, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

