Uncovering molecular biomarkers that correlate cognitive decline with the changes of hippocampus' gene expression profiles in Alzheimer's disease
- PMID: 20405009
- PMCID: PMC2854141
- DOI: 10.1371/journal.pone.0010153
Uncovering molecular biomarkers that correlate cognitive decline with the changes of hippocampus' gene expression profiles in Alzheimer's disease
Abstract
Background: Alzheimer's disease (AD) is characterized by a neurodegenerative progression that alters cognition. On a phenotypical level, cognition is evaluated by means of the MiniMental State Examination (MMSE) and the post-mortem examination of Neurofibrillary Tangle count (NFT) helps to confirm an AD diagnostic. The MMSE evaluates different aspects of cognition including orientation, short-term memory (retention and recall), attention and language. As there is a normal cognitive decline with aging, and death is the final state on which NFT can be counted, the identification of brain gene expression biomarkers from these phenotypical measures has been elusive.
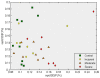
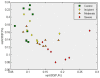
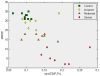
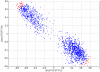
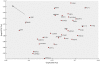
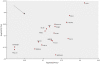
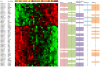
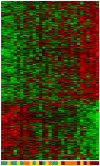
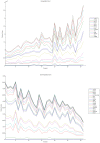
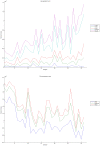
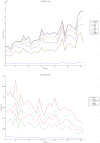
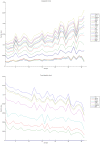
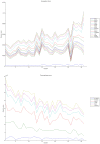
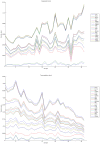
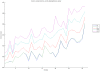
Methodology/principal findings: We have reanalysed a microarray dataset contributed in 2004 by Blalock et al. of 31 samples corresponding to hippocampus gene expression from 22 AD subjects of varying degree of severity and 9 controls. Instead of only relying on correlations of gene expression with the associated MMSE and NFT measures, and by using modern bioinformatics methods based on information theory and combinatorial optimization, we uncovered a 1,372-probe gene expression signature that presents a high-consensus with established markers of progression in AD. The signature reveals alterations in calcium, insulin, phosphatidylinositol and wnt-signalling. Among the most correlated gene probes with AD severity we found those linked to synaptic function, neurofilament bundle assembly and neuronal plasticity.
Conclusions/significance: A transcription factors analysis of 1,372-probe signature reveals significant associations with the EGR/KROX family of proteins, MAZ, and E2F1. The gene homologous of EGR1, zif268, Egr-1 or Zenk, together with other members of the EGR family, are consolidating a key role in the neuronal plasticity in the brain. These results indicate a degree of commonality between putative genes involved in AD and prion-induced neurodegenerative processes that warrants further investigation.
Conflict of interest statement
Figures

References
-
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. - PubMed
-
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, et al. Incipient Alzheimer's disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2173–2178. - PMC - PubMed
-
- Schmitt FA, Davis DG, Wekstein DR, Smith CD, Ashford JW, et al. “Preclinical” AD revisited: Neuropathology of cognitively normal older adults. Neurology. 2000;55:370–376. - PubMed
-
- Haroutunian V, Purohit DP, Perl DP, Marin D, Khan K, et al. Neurofibrillary Tangles in Nondemented Elderly Subjects and Mild Alzheimer Disease. Arch Neurol. 1999;56:713–718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

