Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli
- PMID: 20405029
- PMCID: PMC2853565
- DOI: 10.1371/journal.pone.0010132
Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli
Abstract
Background: Hydrogen production by fermenting bacteria such as Escherichia coli offers a potential source of hydrogen biofuel. Because H(2) production involves consumption of 2H(+), hydrogenase expression is likely to involve pH response and regulation. Hydrogenase consumption of protons in E. coli has been implicated in acid resistance, the ability to survive exposure to acid levels (pH 2-2.5) that are three pH units lower than the pH limit of growth (pH 5-6). Enhanced survival in acid enables a larger infective inoculum to pass through the stomach and colonize the intestine. Most acid resistance mechanisms have been defined using aerobic cultures, but the use of anaerobic cultures will reveal novel acid resistance mechanisms.
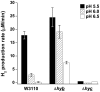
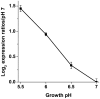
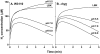
Methods and principal findings: We analyzed the pH regulation of bacterial hydrogenases in live cultures of E. coli K-12 W3110. During anaerobic growth in the range of pH 5 to 6.5, E. coli expresses three hydrogenase isoenzymes that reversibly oxidize H(2) to 2H(+). Anoxic conditions were used to determine which of the hydrogenase complexes contribute to acid resistance, measured as the survival of cultures grown at pH 5.5 without aeration and exposed for 2 hours at pH 2 or at pH 2.5. Survival of all strains in extreme acid was significantly lower in low oxygen than for aerated cultures. Deletion of hyc (Hyd-3) decreased anoxic acid survival 3-fold at pH 2.5, and 20-fold at pH 2, but had no effect on acid survival with aeration. Deletion of hyb (Hyd-2) did not significantly affect acid survival. The pH-dependence of H(2) production and consumption was tested using a H(2)-specific Clark-type electrode. Hyd-3-dependent H(2) production was increased 70-fold from pH 6.5 to 5.5, whereas Hyd-2-dependent H(2) consumption was maximal at alkaline pH. H(2) production, was unaffected by a shift in external or internal pH. H(2) production was associated with hycE expression levels as a function of external pH.
Conclusions: Anaerobic growing cultures of E. coli generate H(2) via Hyd-3 at low external pH, and consume H(2) via Hyd-2 at high external pH. Hyd-3 proton conversion to H(2) is required for acid resistance in anaerobic cultures of E. coli.
Conflict of interest statement
Figures



References
-
- Hansel A, Lindblad P. Towards optimization of cyanobacteria as biotechnologically relevant producers of molecular hydrogen, a clean and renewable energy source. Appl Microbiol Biotechnol. 1998;50:153–160.
-
- Valdez-Vazquez I, Poggi-Varaldo HM. Hydrogen production by fermentative consortia. Renewable Sustainable Energy Rev. 2009;13:1000–1013.
-
- Moss AR, Jouany J-P, Newbold J. Methane production by ruminants: its contribution to global warming. Ann Zootech. 2000;49:231–253.
-
- Olson JW, Maier RJ. Molecular hydrogen as an energy source of Helicobacter pylori. Science. 2002;298:1788–1790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

