Interaction and interrelation of P2X7 and P2X4 receptor complexes in mouse lung epithelial cells
- PMID: 20405163
- PMCID: PMC11115700
- DOI: 10.1007/s00018-010-0355-1
Interaction and interrelation of P2X7 and P2X4 receptor complexes in mouse lung epithelial cells
Abstract
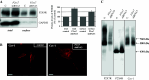
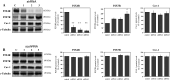
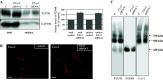
P2X4 and P2X7 receptors are ATP-gated ion channels that are co-expressed in alveolar epithelial type I cells. Both receptors are localized to the plasma membrane and partly associated with lipid rafts. Here we report on our study in an alveolar epithelial cell line of the molecular organization of P2X7R and P2X4R receptors and the effect of their knockdown. Native gel electrophoresis reveals three P2X7R complexes of approximately 430, approximately 580 and approximately 760 kDa. The latter two correspond exactly in size to signals of Cav-1, the structural protein of caveolae. Interestingly knockdown of P2rx7 affects protein levels, the intracellular distribution and the supramolecular organization of Cav-1 as well as of P2X4R, which is mainly detected in a complex of approximately 430 kDa. Our data suggest upregulation of P2X4R as a compensatory mechanism of P2X7R depletion.
Figures




Similar articles
-
Different localization of P2X4 and P2X7 receptors in native mouse lung - lack of evidence for a direct P2X4-P2X7 receptor interaction.Front Immunol. 2024 Jun 17;15:1425938. doi: 10.3389/fimmu.2024.1425938. eCollection 2024. Front Immunol. 2024. PMID: 38953020 Free PMC article.
-
Activation of P2X7R and downstream effects in bleomycin treated lung epithelial cells.Int J Biochem Cell Biol. 2012 Mar;44(3):514-24. doi: 10.1016/j.biocel.2011.12.003. Epub 2011 Dec 14. Int J Biochem Cell Biol. 2012. PMID: 22192844
-
Analysis of assembly and trafficking of native P2X4 and P2X7 receptor complexes in rodent immune cells.J Biol Chem. 2009 May 15;284(20):13446-13454. doi: 10.1074/jbc.M901255200. Epub 2009 Mar 20. J Biol Chem. 2009. PMID: 19304656 Free PMC article.
-
Membrane compartments and purinergic signalling: occurrence and function of P2X receptors in lung.FEBS J. 2009 Jan;276(2):341-53. doi: 10.1111/j.1742-4658.2008.06795.x. Epub 2008 Dec 9. FEBS J. 2009. PMID: 19076210 Review.
-
Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling.Biochim Biophys Acta. 2014 Feb;1838(2):532-45. doi: 10.1016/j.bbamem.2013.07.018. Epub 2013 Jul 27. Biochim Biophys Acta. 2014. PMID: 23899502 Free PMC article. Review.
Cited by
-
P2X7 receptors in body temperature, locomotor activity, and brain mRNA and lncRNA responses to sleep deprivation.Am J Physiol Regul Integr Comp Physiol. 2016 Dec 1;311(6):R1004-R1012. doi: 10.1152/ajpregu.00167.2016. Epub 2016 Oct 5. Am J Physiol Regul Integr Comp Physiol. 2016. PMID: 27707719 Free PMC article.
-
Lack of neuroprotection in the absence of P2X7 receptors in toxin-induced animal models of Parkinson's disease.Mol Neurodegener. 2011 May 4;6:28. doi: 10.1186/1750-1326-6-28. Mol Neurodegener. 2011. PMID: 21542899 Free PMC article.
-
Interaction of Purinergic P2X4 and P2X7 Receptor Subunits.Front Pharmacol. 2017 Nov 22;8:860. doi: 10.3389/fphar.2017.00860. eCollection 2017. Front Pharmacol. 2017. PMID: 29213241 Free PMC article.
-
Detection of caveolin-3/caveolin-1/P2X7R complexes in mice atrial cardiomyocytes in vivo and in vitro.Histochem Cell Biol. 2012 Aug;138(2):231-41. doi: 10.1007/s00418-012-0961-0. Epub 2012 May 15. Histochem Cell Biol. 2012. PMID: 22585038 Free PMC article.
-
P2X4 receptors influence inflammasome activation after spinal cord injury.J Neurosci. 2012 Feb 29;32(9):3058-66. doi: 10.1523/JNEUROSCI.4930-11.2012. J Neurosci. 2012. PMID: 22378878 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

