Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV- 1 maturation inhibitor bevirimat
- PMID: 20406463
- PMCID: PMC2873507
- DOI: 10.1186/1742-4690-7-36
Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV- 1 maturation inhibitor bevirimat
Abstract
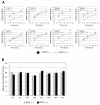
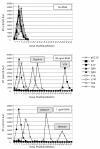
Background: The maturation inhibitor bevirimat (BVM) potently inhibits human immunodeficiency virus type 1 (HIV-1) replication by blocking capsid-spacer peptide 1 (CA-SP1) cleavage. Recent clinical trials demonstrated that a significant proportion of HIV-1-infected patients do not respond to BVM. A patient's failure to respond correlated with baseline polymorphisms at SP1 residues 6-8.
Results: In this study, we demonstrate that varying levels of BVM resistance are associated with point mutations at these residues. BVM susceptibility was maintained by SP1-Q6A, -Q6H and -T8A mutations. However, an SP1-V7A mutation conferred high-level BVM resistance, and SP1-V7M and T8Delta mutations conferred intermediate levels of BVM resistance.
Conclusions: Future exploitation of the CA-SP1 cleavage site as an antiretroviral drug target will need to overcome the baseline variability in the SP1 region of Gag.
Figures

References
-
- Vogt VM. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. - PubMed
-
- Mervis RJ, Ahmad N, Lillehoj EP, Raum MG, Salazar FH, Chan HW, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

