RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis
- PMID: 20406785
- PMCID: PMC2882256
- DOI: 10.1093/jxb/erq088
RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis
Abstract
Although our understanding of mechanisms of DNA repair in bacteria and eukaryotic nuclei continues to improve, almost nothing is known about the DNA repair process in plant organelles, especially chloroplasts. Since the RecA protein functions in DNA repair for bacteria, an analogous function may exist for chloroplasts. The effects on chloroplast DNA (cpDNA) structure of two nuclear-encoded, chloroplast-targeted homologues of RecA in Arabidopsis were examined. A homozygous T-DNA insertion mutation in one of these genes (cpRecA) resulted in altered structural forms of cpDNA molecules and a reduced amount of cpDNA, while a similar mutation in the other gene (DRT100) had no effect. Double mutants exhibited a similar phenotype to cprecA single mutants. The cprecA mutants also exhibited an increased amount of single-stranded cpDNA, consistent with impaired RecA function. After four generations, the cprecA mutant plants showed signs of reduced chloroplast function: variegation and necrosis. Double-stranded breaks in cpDNA of wild-type plants caused by ciprofloxacin (an inhibitor of Escherichia coli gyrase, a type II topoisomerase) led to an alteration of cpDNA structure that was similar to that seen in cprecA mutants. It is concluded that the process by which damaged DNA is repaired in bacteria has been retained in their endosymbiotic descendent, the chloroplast.
Figures

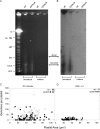
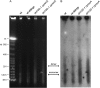
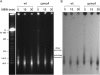
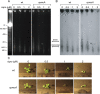

Comment in
-
Mechanisms for maintenance, replication, and repair of the chloroplast genome in plants.J Exp Bot. 2010 Jun;61(10):2535-7. doi: 10.1093/jxb/erq163. Epub 2010 May 30. J Exp Bot. 2010. PMID: 20513654 No abstract available.
References
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nature Reviews Genetics. 2008;9:204–217. - PubMed
-
- Backert S, Dorfel P, Börner T. Investigation of plant organellar DNAs by pulsed-field gel electrophoresis. Current Genetics. 1995;28:390–399. - PubMed
-
- Bendich AJ, Smith SB. Moving pictures and pulsed-field gel electrophoresis show linear DNA molecules from chloroplasts and mitochondria. Current Genetics. 1990;17:421–425. - PubMed
-
- Birky CW., Jr The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annual Review of Genetics. 2001;35:125–148. - PubMed
-
- Bohr VA, Anson RM. Mitochondrial DNA repair pathways. Journal of Bioenergetics and Biomembranes. 1999;31:391–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

