Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets
- PMID: 20406816
- PMCID: PMC2885207
- DOI: 10.1074/jbc.M110.134213
Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets
Abstract
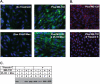
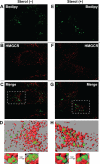
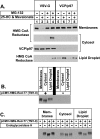
Sterol-induced binding to Insigs in the endoplasmic reticulum (ER) allows for ubiquitination of 3-hydroxy-3-methylglutaryl coenzyme A reductase, the rate-limiting enzyme in cholesterol synthesis. This ubiquitination marks reductase for recognition by the ATPase VCP/p97, which mediates extraction and delivery of reductase from ER membranes to cytosolic 26 S proteasomes for degradation. Here, we report that reductase becomes dislocated from ER membranes into the cytosol of sterol-treated cells. This dislocation exhibits an absolute requirement for the actions of Insigs and VCP/p97. Reductase also appears in a buoyant fraction of sterol-treated cells that co-purifies with lipid droplets, cytosolic organelles traditionally regarded as storage depots for neutral lipids such as triglycerides and cholesteryl esters. Genetic, biochemical, and localization studies suggest a model in which reductase is dislodged into the cytosol from an ER subdomain closely associated with lipid droplets.
Figures



References
-
- Brown M. S., Goldstein J. L. (1980) J. Lipid Res 21, 505–517 - PubMed
-
- Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. (1985) J. Biol. Chem. 260, 522–530 - PubMed
-
- Nakanishi M., Goldstein J. L., Brown M. S. (1988) J. Biol. Chem. 263, 8929–8937 - PubMed
-
- Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. (2003) Mol. Cell 11, 25–33 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

