Chromosomal and microRNA expression patterns reveal biologically distinct subgroups of 11q- neuroblastoma
- PMID: 20406844
- PMCID: PMC2880207
- DOI: 10.1158/1078-0432.CCR-09-3215
Chromosomal and microRNA expression patterns reveal biologically distinct subgroups of 11q- neuroblastoma
Abstract
Purpose: The purpose of this study was to further define the biology of the 11q- neuroblastoma tumor subgroup by the integration of array-based comparative genomic hybridization with microRNA (miRNA) expression profiling data to determine if improved patient stratification is possible.
Experimental design: A set of primary neuroblastoma (n = 160), which was broadly representative of all genetic subtypes, was analyzed by array-based comparative genomic hybridization and for the expression of 430 miRNAs. A 15-miRNA expression signature previously shown to be predictive of clinical outcome was used to analyze an independent cohort of 11q- tumors (n = 37).
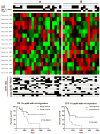
Results: Loss of 4p and gain of 7q occurred at a significantly higher frequency in the 11q- tumors, further defining the genetic characteristics of this subtype. The 11q- tumors could be split into two subgroups using a miRNA expression survival signature that differed significantly in clinical outcome and the overall frequency of large-scale genomic imbalances, with the poor survival subgroup having significantly more imbalances. miRNAs from the expression signature, which were upregulated in unfavorable tumors, were predicted to target downregulated genes from a published mRNA expression classifier of clinical outcome at a higher-than-expected frequency, indicating the miRNAs might contribute to the regulation of genes within the signature.
Conclusion: We show that two distinct biological subtypes of neuroblastoma with loss of 11q occur, which differ in their miRNA expression profiles, frequency of segmental imbalances, and clinical outcome. A miRNA expression signature, combined with an analysis of segmental imbalances, provides greater prediction of event-free survival and overall survival outcomes than 11q status by itself, improving patient stratification.
Copyright 2010 AACR.
Figures

Comment in
-
Pediatric oncology: 11q negative neuroblastoma subgroups predict outcomes and may guide treatment.Nat Rev Clin Oncol. 2010 Aug;7(8):421. doi: 10.1038/nrclinonc.2010.110. Nat Rev Clin Oncol. 2010. PMID: 20700946 No abstract available.
References
-
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. - PubMed
-
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. - PubMed
-
- Van Roy N, Laureys G, Cheng NC, et al. 1;17 translocations and other chromosome 17 rearrangements in human primary neuroblastoma tumors and cell lines. Genes Chromosomes Cancer. 1994;10:103–14. - PubMed
-
- Guo C, White PS, Weiss MJ, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18:4948–57. - PubMed
-
- Plantaz D, Vandesompele J, Van Roy N, et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer. 2001;91:680–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

