A variant of the KLK4 gene is expressed as a cis sense-antisense chimeric transcript in prostate cancer cells
- PMID: 20406994
- PMCID: PMC2874168
- DOI: 10.1261/rna.2019810
A variant of the KLK4 gene is expressed as a cis sense-antisense chimeric transcript in prostate cancer cells
Abstract
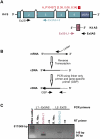
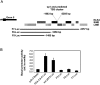
In humans, more than 30,000 chimeric transcripts originating from 23,686 genes have been identified. The mechanisms and association of chimeric transcripts arising from chromosomal rearrangements with cancer are well established, but much remains unknown regarding the biogenesis and importance of other chimeric transcripts that arise from nongenomic alterations. Recently, a SLC45A3-ELK4 chimera has been shown to be androgen-regulated, and is overexpressed in metastatic or high-grade prostate tumors relative to local prostate cancers. Here, we characterize the expression of a KLK4 cis sense-antisense chimeric transcript, and show other examples in prostate cancer. Using non-protein-coding microarray analyses, we initially identified an androgen-regulated antisense transcript within the 3' untranslated region of the KLK4 gene in LNCaP cells. The KLK4 cis-NAT was validated by strand-specific linker-mediated RT-PCR and Northern blotting. Characterization of the KLK4 cis-NAT by 5' and 3' rapid amplification of cDNA ends (RACE) revealed that this transcript forms multiple fusions with the KLK4 sense transcript. Lack of KLK4 antisense promoter activity using reporter assays suggests that these transcripts are unlikely to arise from a trans-splicing mechanism. 5' RACE and analyses of deep sequencing data from LNCaP cells treated +/-androgens revealed six high-confidence sense-antisense chimeras of which three were supported by the cDNA databases. In this study, we have shown complex gene expression at the KLK4 locus that might be a hallmark of cis sense-antisense chimeric transcription.
Figures




Similar articles
-
Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation.Cancer Discov. 2012 Jul;2(7):598-607. doi: 10.1158/2159-8290.CD-12-0042. Epub 2012 Jun 19. Cancer Discov. 2012. PMID: 22719019
-
SLC45A3-ELK4 chimera in prostate cancer: spotlight on cis-splicing.Cancer Discov. 2012 Jul;2(7):582-5. doi: 10.1158/2159-8290.CD-12-0212. Cancer Discov. 2012. PMID: 22787087 Free PMC article.
-
SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer.Cancer Res. 2009 Apr 1;69(7):2734-8. doi: 10.1158/0008-5472.CAN-08-4926. Epub 2009 Mar 17. Cancer Res. 2009. PMID: 19293179 Free PMC article.
-
Involvement of the multiple tumor suppressor genes and 12-lipoxygenase in human prostate cancer. Therapeutic implications.Adv Exp Med Biol. 1997;407:41-53. doi: 10.1007/978-1-4899-1813-0_7. Adv Exp Med Biol. 1997. PMID: 9321930 Review.
-
Chimeric RNAs in cancer.Adv Clin Chem. 2021;100:1-35. doi: 10.1016/bs.acc.2020.04.001. Epub 2020 May 27. Adv Clin Chem. 2021. PMID: 33453863 Review.
Cited by
-
Pan-Cancer Analysis Reveals the Diverse Landscape of Novel Sense and Antisense Fusion Transcripts.Mol Ther Nucleic Acids. 2020 Mar 6;19:1379-1398. doi: 10.1016/j.omtn.2020.01.023. Epub 2020 Jan 29. Mol Ther Nucleic Acids. 2020. PMID: 32160708 Free PMC article.
-
A microsatellite repeat in PCA3 long non-coding RNA is associated with prostate cancer risk and aggressiveness.Sci Rep. 2017 Dec 4;7(1):16862. doi: 10.1038/s41598-017-16700-y. Sci Rep. 2017. PMID: 29203868 Free PMC article.
-
Strategies to Study the Functions of Pseudogenes in Mouse Models of Cancer.Methods Mol Biol. 2021;2324:287-304. doi: 10.1007/978-1-0716-1503-4_18. Methods Mol Biol. 2021. PMID: 34165722 Review.
-
Pseudogene Associated Recurrent Gene Fusion in Prostate Cancer.Neoplasia. 2019 Oct;21(10):989-1002. doi: 10.1016/j.neo.2019.07.010. Epub 2019 Aug 22. Neoplasia. 2019. PMID: 31446281 Free PMC article.
-
KLK4T2 Is a Hormonally Regulated Transcript from the KLK4 Locus.Int J Mol Sci. 2021 Dec 1;22(23):13023. doi: 10.3390/ijms222313023. Int J Mol Sci. 2021. PMID: 34884832 Free PMC article.
References
-
- Cocquet J, Chong A, Zhang G, Veitia RA 2006. Reverse transcriptase template switching and false alternative transcripts. Genomics 88: 127–131 - PubMed
-
- Das HK, Jackson CL, Miller DA, Leff T, Breslow JL 1987. The human apolipoprotein C-II gene sequence contains a novel chromosome 19-specific minisatellite in its third intron. J Biol Chem 262: 4787–4793 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
