Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network
- PMID: 20407024
- PMCID: PMC2879739
- DOI: 10.1105/tpc.109.073635
Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network
Abstract
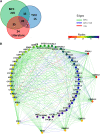
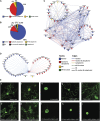
As in other eukaryotes, cell division in plants is highly conserved and regulated by cyclin-dependent kinases (CDKs) that are themselves predominantly regulated at the posttranscriptional level by their association with proteins such as cyclins. Although over the last years the knowledge of the plant cell cycle has considerably increased, little is known on the assembly and regulation of the different CDK complexes. To map protein-protein interactions between core cell cycle proteins of Arabidopsis thaliana, a binary protein-protein interactome network was generated using two complementary high-throughput interaction assays, yeast two-hybrid and bimolecular fluorescence complementation. Pairwise interactions among 58 core cell cycle proteins were tested, resulting in 357 interactions, of which 293 have not been reported before. Integration of the binary interaction results with cell cycle phase-dependent expression information and localization data allowed the construction of a dynamic interaction network. The obtained interaction map constitutes a framework for further in-depth analysis of the cell cycle machinery.
Figures




References
-
- Akada R., Yamamoto J., Yamashita I. (1997). Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254: 267–274 - PubMed
-
- Aoki K., Ogata Y., Shibata D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48: 381–390 - PubMed
-
- Boniotti M.B., Gutierrez C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28: 341–350 - PubMed
-
- Boudolf V., Vlieghe K., Beemster G.T.S., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

