Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations
- PMID: 20407129
- PMCID: PMC2907208
- DOI: 10.1534/genetics.110.116459
Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations
Abstract
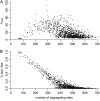
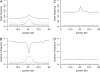
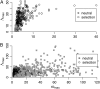
A major goal of population genomics is to reconstruct the history of natural populations and to infer the neutral and selective scenarios that can explain the present-day polymorphism patterns. However, the separation between neutral and selective hypotheses has proven hard, mainly because both may predict similar patterns in the genome. This study focuses on the development of methods that can be used to distinguish neutral from selective hypotheses in equilibrium and nonequilibrium populations. These methods utilize a combination of statistics on the basis of the site frequency spectrum (SFS) and linkage disequilibrium (LD). We investigate the patterns of genetic variation along recombining chromosomes using a multitude of comparisons between neutral and selective hypotheses, such as selection or neutrality in equilibrium and nonequilibrium populations and recurrent selection models. We perform hypothesis testing using the classical P-value approach, but we also introduce methods from the machine-learning field. We demonstrate that the combination of SFS- and LD-based statistics increases the power to detect recent positive selection in populations that have experienced past demographic changes.
Figures
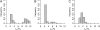
Similar articles
-
On the utility of linkage disequilibrium as a statistic for identifying targets of positive selection in nonequilibrium populations.Genetics. 2007 Aug;176(4):2371-9. doi: 10.1534/genetics.106.069450. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565955 Free PMC article.
-
Detecting Positive Selection in Populations Using Genetic Data.Methods Mol Biol. 2020;2090:87-123. doi: 10.1007/978-1-0716-0199-0_5. Methods Mol Biol. 2020. PMID: 31975165
-
A population genetic hidden Markov model for detecting genomic regions under selection.Mol Biol Evol. 2010 Jul;27(7):1673-85. doi: 10.1093/molbev/msq053. Epub 2010 Feb 25. Mol Biol Evol. 2010. PMID: 20185453 Free PMC article.
-
A survey of methods and tools to detect recent and strong positive selection.J Biol Res (Thessalon). 2017 Apr 8;24:7. doi: 10.1186/s40709-017-0064-0. eCollection 2017 Dec. J Biol Res (Thessalon). 2017. PMID: 28405579 Free PMC article. Review.
-
Genome-wide variation in the human and fruitfly: a comparison.Curr Opin Genet Dev. 2001 Dec;11(6):627-34. doi: 10.1016/s0959-437x(00)00245-8. Curr Opin Genet Dev. 2001. PMID: 11682305 Review.
Cited by
-
The Impact of Genetic Surfing on Neutral Genomic Diversity.Mol Biol Evol. 2022 Nov 3;39(11):msac249. doi: 10.1093/molbev/msac249. Mol Biol Evol. 2022. PMID: 36403964 Free PMC article.
-
Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population.Genetics. 2013 Jan;193(1):291-301. doi: 10.1534/genetics.112.145912. Epub 2012 Nov 12. Genetics. 2013. PMID: 23150605 Free PMC article.
-
Discovery of Ongoing Selective Sweeps within Anopheles Mosquito Populations Using Deep Learning.Mol Biol Evol. 2021 Mar 9;38(3):1168-1183. doi: 10.1093/molbev/msaa259. Mol Biol Evol. 2021. PMID: 33022051 Free PMC article.
-
Soft shoulders ahead: spurious signatures of soft and partial selective sweeps result from linked hard sweeps.Genetics. 2015 May;200(1):267-84. doi: 10.1534/genetics.115.174912. Epub 2015 Feb 25. Genetics. 2015. PMID: 25716978 Free PMC article.
-
Divergent Selection and Primary Gene Flow Shape Incipient Speciation of a Riparian Tree on Hawaii Island.Mol Biol Evol. 2020 Mar 1;37(3):695-710. doi: 10.1093/molbev/msz259. Mol Biol Evol. 2020. PMID: 31693149 Free PMC article.
References
-
- Barton, N., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72 123–133.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

