Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion
- PMID: 20407419
- PMCID: PMC2876962
- DOI: 10.1038/emboj.2010.54
Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion
Abstract
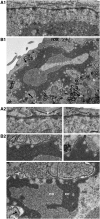
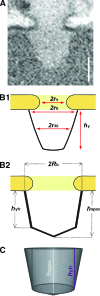
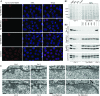
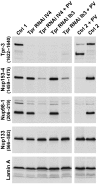
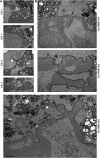
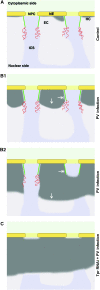
Amassments of heterochromatin in somatic cells occur in close contact with the nuclear envelope (NE) but are gapped by channel- and cone-like zones that appear largely free of heterochromatin and associated with the nuclear pore complexes (NPCs). To identify proteins involved in forming such heterochromatin exclusion zones (HEZs), we used a cell culture model in which chromatin condensation induced by poliovirus (PV) infection revealed HEZs resembling those in normal tissue cells. HEZ occurrence depended on the NPC-associated protein Tpr and its large coiled coil-forming domain. RNAi-mediated loss of Tpr allowed condensing chromatin to occur all along the NE's nuclear surface, resulting in HEZs no longer being established and NPCs covered by heterochromatin. These results assign a central function to Tpr as a determinant of perinuclear organization, with a direct role in forming a morphologically distinct nuclear sub-compartment and delimiting heterochromatin distribution.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O (2004) Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306: 1387–1390 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

