Statistical contributions to proteomic research
- PMID: 20407946
- PMCID: PMC3889133
- DOI: 10.1007/978-1-60761-711-2_9
Statistical contributions to proteomic research
Abstract
Proteomic profiling has the potential to impact the diagnosis, prognosis, and treatment of various diseases. A number of different proteomic technologies are available that allow us to look at many proteins at once, and all of them yield complex data that raise significant quantitative challenges. Inadequate attention to these quantitative issues can prevent these studies from achieving their desired goals, and can even lead to invalid results. In this chapter, we describe various ways the involvement of statisticians or other quantitative scientists in the study team can contribute to the success of proteomic research, and we outline some of the key statistical principles that should guide the experimental design and analysis of such studies.
Figures


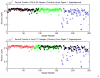
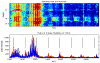
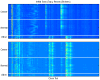


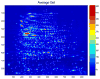
References
-
- Baggerly KA, Morris JS, Wang J, Gold D, Xiao LC, Coombes KR. A comprehensive approach to the analysis of matrix-assisted laser desorption/ionization time of flight proteomics spectra from serum samples. Proteomics. 2003;3:1667–1672. - PubMed
-
- Baggerly KA, Edmonson S, Morris JS, Coombes KR. High-Resolution Serum Proteomic Patterns for Ovarian Cancer Detection. Endocrine-Related Cancers. 2004;11(4):583–584. - PubMed
-
- Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. - PubMed
-
- Baggerly KA, Morris JS, Edmonson S, Coombes KR. Signal in Noise: Evaluating Reported Reproducibility of Serum Proteomic Tests for Ovarian Cancer. Journal of the National Cancer Institute. 2005;97:307–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

