MHC class II epitope predictive algorithms
- PMID: 20408898
- PMCID: PMC2913211
- DOI: 10.1111/j.1365-2567.2010.03268.x
MHC class II epitope predictive algorithms
Abstract
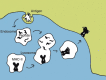
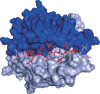
Major histocompatibility complex class II (MHC-II) molecules sample peptides from the extracellular space, allowing the immune system to detect the presence of foreign microbes from this compartment. To be able to predict the immune response to given pathogens, a number of methods have been developed to predict peptide-MHC binding. However, few methods other than the pioneering TEPITOPE/ProPred method have been developed for MHC-II. Despite recent progress in method development, the predictive performance for MHC-II remains significantly lower than what can be obtained for MHC-I. One reason for this is that the MHC-II molecule is open at both ends allowing binding of peptides extending out of the groove. The binding core of MHC-II-bound peptides is therefore not known a priori and the binding motif is hence not readily discernible. Recent progress has been obtained by including the flanking residues in the predictions. All attempts to make ab initio predictions based on protein structure have failed to reach predictive performances similar to those that can be obtained by data-driven methods. Thousands of different MHC-II alleles exist in humans. Recently developed pan-specific methods have been able to make reasonably accurate predictions for alleles that were not included in the training data. These methods can be used to define supertypes (clusters) of MHC-II alleles where alleles within each supertype have similar binding specificities. Furthermore, the pan-specific methods have been used to make a graphical atlas such as the MHCMotifviewer, which allows for visual comparison of specificities of different alleles.
Figures
References
-
- Castellino F, Zhong G, Germain RN. Antigen presentation by MHC class II molecules: invariant chain function, protein trafficking, and the molecular basis of diverse determinant capture. Hum Immunol. 1997;54:159–69. - PubMed
-
- Mouritsen S, Meldal M, Werdelin O, Hansen AS, Buus S. MHC molecules protect T cell epitopes against proteolytic destruction. J Immunol. 1992;149:1987–93. - PubMed
-
- Chapman HA. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. - PubMed
-
- Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol. 2004;5:685–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

