Predictors of natively unfolded proteins: unanimous consensus score to detect a twilight zone between order and disorder in generic datasets
- PMID: 20409339
- PMCID: PMC2877690
- DOI: 10.1186/1471-2105-11-198
Predictors of natively unfolded proteins: unanimous consensus score to detect a twilight zone between order and disorder in generic datasets
Abstract
Background: Natively unfolded proteins lack a well defined three dimensional structure but have important biological functions, suggesting a re-assignment of the structure-function paradigm. To assess that a given protein is natively unfolded requires laborious experimental investigations, then reliable sequence-only methods for predicting whether a sequence corresponds to a folded or to an unfolded protein are of interest in fundamental and applicative studies. Many proteins have amino acidic compositions compatible both with the folded and unfolded status, and belong to a twilight zone between order and disorder. This makes difficult a dichotomic classification of protein sequences into folded and natively unfolded ones. In this work we propose an operational method to identify proteins belonging to the twilight zone by combining into a consensus score good performing single predictors of folding.
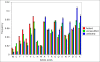
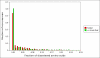
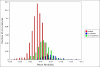
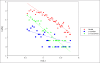
Results: In this methodological paper dichotomic folding indexes are considered: hydrophobicity-charge, mean packing, mean pairwise energy, Poodle-W and a new global index, that is called here gVSL2, based on the local disorder predictor VSL2. The performance of these indexes is evaluated on different datasets, in particular on a new dataset composed by 2369 folded and 81 natively unfolded proteins. Poodle-W, gVSL2 and mean pairwise energy have good performance and stability in all the datasets considered and are combined into a strictly unanimous combination score SSU, that leaves proteins unclassified when the consensus of all combined indexes is not reached. The unclassified proteins: i) belong to an overlap region in the vector space of amino acidic compositions occupied by both folded and unfolded proteins; ii) are composed by approximately the same number of order-promoting and disorder-promoting amino acids; iii) have a mean flexibility intermediate between that of folded and that of unfolded proteins.
Conclusions: Our results show that proteins unclassified by SSU belong to a twilight zone. Proteins left unclassified by the consensus score SSU have physical properties intermediate between those of folded and those of natively unfolded proteins and their structural properties and evolutionary history are worth to be investigated.
Figures




References
-
- Dunker A, Lawson J, Brown C, Romero P, Oh J, Oldfield C, Campen A, Ratliffl C, Hipps K, Ausio J, Nissen M, Reeves R, Kang C, Kissinger C, Bailey R, Griswold M, Chin W, Garner E, Obradovic Z. Intrinsically disordered proteins. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/S1093-3263(00)00138-8. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

