Force transduction by the microtubule-bound Dam1 ring
- PMID: 20409480
- PMCID: PMC2856180
- DOI: 10.1016/j.bpj.2010.01.004
Force transduction by the microtubule-bound Dam1 ring
Abstract
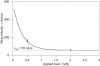
The coupling between the depolymerization of microtubules (MTs) and the motion of the Dam1 ring complex is now thought to play an important role in the generation of forces during mitosis. Our current understanding of this motion is based on a number of detailed computational models. Although these models realize possible mechanisms for force transduction, they can be extended by variation of any of a large number of poorly measured parameters and there is no clear strategy for determining how they might be distinguished experimentally. Here we seek to identify and analyze two distinct mechanisms present in the computational models. In the first, the splayed protofilaments at the end of the depolymerizing MT physically prevent the Dam1 ring from falling off the end, and in the other, an attractive binding secures the ring to the microtubule. Based on this analysis, we discuss how to distinguish between competing models that seek to explain how the Dam1 ring stays on the MT. We propose novel experimental approaches that could resolve these models for the first time, either by changing the diffusion constant of the Dam1 ring (e.g., by tethering a long polymer to it) or by using a time-varying load.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


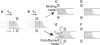

References
-
- Alberts B., Johnson A., Walter P. 4th Ed. Garland Science; New York: 2002. Molecular Biology of the Cell.
-
- Ganem N.J., Upton K., Compton D.A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15:1827–1832. - PubMed
-
- Koshland D.E., Mitchison T.J., Kirschner M.W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

