Predicting secondary structural folding kinetics for nucleic acids
- PMID: 20409482
- PMCID: PMC2856163
- DOI: 10.1016/j.bpj.2009.12.4319
Predicting secondary structural folding kinetics for nucleic acids
Abstract
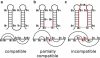
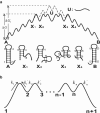
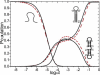
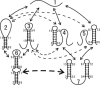
We report a new computational approach to the prediction of RNA secondary structure folding kinetics. In this approach, each elementary kinetic step is represented as the transformation between two secondary structures that differ by a helix. Based on the free energy landscape analysis, we identify three types of dominant pathways and the rate constants for the kinetic steps: 1), formation; 2), disruption of a helix stem; and 3), helix formation with concomitant partial melting of a competing (incompatible) helix. The third pathway, termed the tunneling pathway, is the low-barrier dominant pathway for the conversion between two incompatible helices. Comparisons with experimental data indicate that this new method is quite reliable in predicting the kinetics for RNA secondary structural folding and structural rearrangements. The approach presented here may provide a robust first step for further systematic development of a predictive theory for the folding kinetics for large RNAs.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
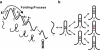
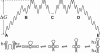
References
-
- Micura R., Höbartner C. On secondary structure rearrangements and equilibria of small RNAs. ChemBioChem. 2003;4:984–990. - PubMed
-
- Fürtig B., Buck J., Schwalbe H. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;86:360–383. - PubMed
-
- Harlepp S., Marchal T., Chatenay D. Probing complex RNA structures by mechanical force. Eur Phys. J. E. Soft Matter. 2003;12:605–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

