The behavior of the hydrophobic effect under pressure and protein denaturation
- PMID: 20409483
- PMCID: PMC2856145
- DOI: 10.1016/j.bpj.2009.12.4298
The behavior of the hydrophobic effect under pressure and protein denaturation
Abstract
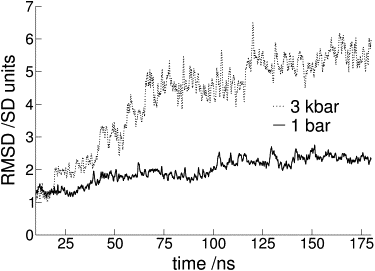
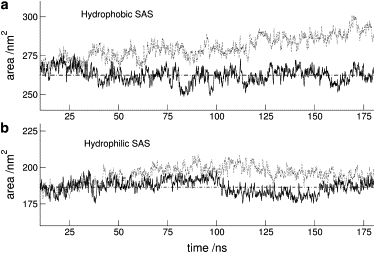
It is well known that proteins denature under high pressure. The mechanism that underlies such a process is still not clearly understood, however, giving way to controversial interpretations. Using molecular dynamics simulation on systems that may be regarded experimentally as limiting examples of the effect of high pressure on globular proteins, such as lysozyme and apomyoglobin, we have effectively reproduced such similarities and differences in behavior as are interpreted from experiment. From the analysis of such data, we explain the experimental evidence at hand through the effect of pressure on the change of water structure, and hence the weakening of the hydrophobic effect that is known to be the main driving force in protein folding.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Kauzmann W. Some factors in the interpretation of the protein denaturation. Adv. Protein Chem. 1959;14:1–63. - PubMed
-
- Jaenicke R. Protein folding: local structures, domains, subunits, and assemblies. Biochemistry. 1991;30:3147–3161. - PubMed
-
- Royer C. Revisiting volume changes in pressure-induced protein unfolding. Biochim Biophys Acta. 2002;1595:201–209. - PubMed
-
- Goldammer V., Hertz H.G. Molecular motion and structure of aqueous mixtures with nonelectrolytes as studied by nuclear magnetic relaxation methods. J. Phys. Chem. 1970;74:3734–3755.
-
- Hallenga K., Grigera J.R., Berendsen H.J.C. Influence of hydrophobic solutes on the dynamic behavior of water. J. Phys. Chem. 1980;84:2381–2390.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

