Protein collective motions coupled to ligand migration in myoglobin
- PMID: 20409486
- PMCID: PMC2856161
- DOI: 10.1016/j.bpj.2009.12.4318
Protein collective motions coupled to ligand migration in myoglobin
Abstract
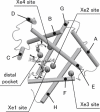
Ligand migration processes inside myoglobin and protein dynamics coupled to the migration were theoretically investigated with molecular dynamics simulations. Based on a linear response theory, we identified protein motions coupled to the transient migration of ligand, carbon monoxide (CO), through channels. The result indicates that the coupled protein motions involve collective motions extended over the entire protein correlated with local gating motions at the channels. Protein motions, coupled to opening of a channel from the distal pocket to a neighboring xenon site, were found to share the collective motion with experimentally observed protein motions coupled to a doming motion of the heme Fe atom upon photodissociation of the ligand. Analysis based on generalized Langevin dynamics elucidated slow and diffusive features of the protein response motions. Remarkably small transmission coefficients for rates of the CO migrations through myoglobin were found, suggesting that the CO migration dynamics are characterized as motions governed by the protein dynamics involving the collective motions, rather than as thermally activated transitions across energy barriers of well-structured channels.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Tilton R.F., Kuntz I.D., Petsko G.A. Cavities in proteins: structure of a metmyoglobin-xenon complex solved to 1.9 Å. Biochemistry. 1984;23:2849–2857. - PubMed
-
- Sakakura M., Yamaguchi S., Terazima M. Dynamics of structure and energy of horse carboxymyoglobin after photodissociation of carbon monoxide. J. Am. Chem. Soc. 2001;123:4286–4294. - PubMed
-
- Chu K., Vojtchovský J., Schlichting I. Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature. 2000;403:921–923. - PubMed
-
- Ostermann A., Waschipky R., Nienhaus G.U. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

