The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated
- PMID: 20409569
- PMCID: PMC7119183
- DOI: 10.1016/j.virol.2010.03.015
The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated
Abstract
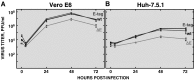
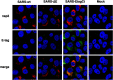
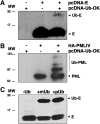
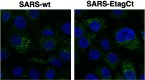
To analyze the proteins interacting with the severe acute respiratory syndrome coronavirus (SARS-CoV) envelope (E) protein, a SARS-CoV was engineered including two tags associated to the E protein. Using this virus, complexes of SARS-CoV E and other proteins were purified using a tandem affinity purification system. Several viral and cell proteins including spike, membrane, non-structural protein 3 (nsp3), dynein heavy chain, fatty acid synthase and transmembrane protein 43 bound E protein. In the present work, we focused on the binding of E protein to nsp3 in infected cells and cell-free systems. This interaction was mediated by the N-terminal acidic domain of nsp3. Moreover, nsp3 and E protein colocalized during the infection. It was shown that E protein was ubiquitinated in vitro and in cell culture, suggesting that the interaction between nsp3 and E protein may play a role in the E protein ubiquitination status and therefore on its turnover.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures




References
-
- de Groot R.J., Ziebuhr J., Poon L.L., Woo P.C., Talbot P., Rottier P.J.M., Holmes K.V., Baric R., Perlman S., Enjuanes L., Gorbalenya A.E. International Committee on Taxonomy of Viruses, 2008.085-126V. 2008. Revision of the family Coronaviridae.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

