Dynamic simulation of cardiolipin remodeling: greasing the wheels for an interpretative approach to lipidomics
- PMID: 20410019
- PMCID: PMC2903823
- DOI: 10.1194/jlr.M004796
Dynamic simulation of cardiolipin remodeling: greasing the wheels for an interpretative approach to lipidomics
Abstract
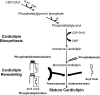
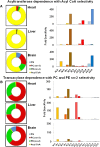
Cardiolipin is a class of mitochondrial specific phospholipid, which is intricately involved in mitochondrial functionality. Differences in cardiolipin species exist in a variety of tissues and diseases. It has been demonstrated that the cardiolipin profile is a key modulator of the functions of many mitochondrial proteins. However, the chemical mechanism(s) leading to normal and/or pathological distribution of cardiolipin species remain elusive. Herein, we describe a novel approach for investigating the molecular mechanism of cardiolipin remodeling through a dynamic simulation. This approach applied data from shotgun lipidomic analyses of the heart, liver, brain, and lung mitochondrial lipidomes to model cardiolipin remodeling, including relative content, regiospecificity, and isomeric composition of cardiolipin species. Generated cardiolipin profiles were nearly identical to those determined by shotgun lipidomics. Importantly, the simulated isomeric compositions of cardiolipin species were further substantiated through product ion analysis. Finally, unique enzymatic activities involved in cardiolipin remodeling were assessed from the parameters used in the dynamic simulation of cardiolipin profiles. Collectively, we described, verified, and demonstrated a novel approach by integrating both lipidomic analysis and dynamic simulation to study cardiolipin biology. We believe this study provides a foundation to investigate cardiolipin metabolism and bioenergetic homeostasis in normal and disease states.
Figures





References
-
- Han X., Gross R. W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. - PubMed
-
- Niemela P. S., Castillo S., Sysi-Aho M., Oresic M. 2009. Bioinformatics and computational methods for lipidomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2855–2862. - PubMed
-
- Han X., Gross R. W. 2005. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev. Proteomics. 2: 253–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

