Dual-targeting siRNAs
- PMID: 20410240
- PMCID: PMC2874179
- DOI: 10.1261/rna.2005710
Dual-targeting siRNAs
Abstract
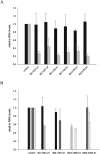
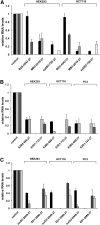
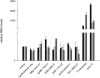
We have developed an algorithm for the prediction of dual-targeting short interfering RNAs (siRNAs) in which both strands are deliberately designed to separately target different mRNA transcripts with complete complementarity. An advantage of this approach versus the use of two separate duplexes is that only two strands, as opposed to four, are competing for entry into the RNA-induced silencing complex. We chose to design our dual-targeting siRNAs as Dicer substrate 25/27mer siRNAs, since design features resembling pre-microRNAs (miRNAs) can be introduced for Dicer processing. Seven different dual-targeting siRNAs targeting genes that are potential targets in cancer therapy have been developed including Bcl2, Stat3, CCND1, BIRC5, and MYC. The dual-targeting siRNAs have been characterized for dual target knockdown in three different cell lines (HEK293, HCT116, and PC3), where they were as effective as their corresponding single-targeting siRNAs in target knockdown. The algorithm developed in this study should prove to be useful for predicting dual-targeting siRNAs in a variety of different targets and is available from http://demo1.interagon.com/DualTargeting/.
Figures


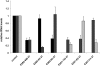
References
-
- Behlke MA 2008. Chemical modification of siRNAs for in vivo use. Oligonucleotides 18: 305–319 - PubMed
-
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3: 199–204 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
