Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo
- PMID: 20410501
- PMCID: PMC2891985
- DOI: 10.1056/NEJMoa0908547
Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo
Abstract
Background: Generalized vitiligo is an autoimmune disease characterized by melanocyte loss, which results in patchy depigmentation of skin and hair, and is associated with an elevated risk of other autoimmune diseases.
Methods: To identify generalized vitiligo susceptibility loci, we conducted a genomewide association study. We genotyped 579,146 single-nucleotide polymorphisms (SNPs) in 1514 patients with generalized vitiligo who were of European-derived white (CEU) ancestry and compared the genotypes with publicly available control genotypes from 2813 CEU persons. We then tested 50 SNPs in two replication sets, one comprising 677 independent CEU patients and 1106 CEU controls and the other comprising 183 CEU simplex trios with generalized vitiligo and 332 CEU multiplex families.
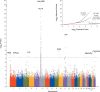
Results: We detected significant associations between generalized vitiligo and SNPs at several loci previously associated with other autoimmune diseases. These included genes encoding major-histocompatibility-complex class I molecules (P=9.05x10(-23)) and class II molecules (P=4.50x10(-34)), PTPN22 (P=1.31x10(-7)), LPP (P=1.01x10(-11)), IL2RA (P=2.78x10(-9)), UBASH3A (P=1.26x10(-9)), and C1QTNF6 (P=2.21x10(-16)). We also detected associations between generalized vitiligo and SNPs in two additional immune-related loci, RERE (P=7.07x10(-15)) and GZMB (P=3.44x10(-8)), and in a locus containing TYR (P=1.60x10(-18)), encoding tyrosinase.
Conclusions: We observed associations between generalized vitiligo and markers implicating multiple genes, some associated with other autoimmune diseases and one (TYR) that may mediate target-cell specificity and indicate a mutually exclusive relationship between susceptibility to vitiligo and susceptibility to melanoma.
2010 Massachusetts Medical Society
Figures

References
-
- Hann S-K, Nordlund JJ. Vitiligo: a monograph on the basic and clinical science. Blackwell Science; Oxford, England: 2000.
-
- Rezaei N, Gavalas NG, Weetman AP, Kemp EH. Autoimmunity as an aetiological factor in vitiligo. J Eur Acad Dermatol Venereol. 2007;21:865–76. - PubMed
-
- Spritz RA. The genetics of generalized vitiligo. Curr Dir Autoimmun. 2008;10:244–57. - PubMed
-
- Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–14. - PubMed
-
- Taïeb A, Picardo M. The definition and assessment of vitiligo: a consensus report of the Vitiligo European Task Force. Pigment Cell Res. 2007;20:27–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
