Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate
- PMID: 20410882
- PMCID: PMC2967782
- DOI: 10.1038/nature08977
Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate
Abstract
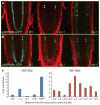
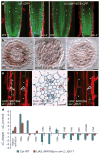
A key question in developmental biology is how cells exchange positional information for proper patterning during organ development. In plant roots the radial tissue organization is highly conserved with a central vascular cylinder in which two water conducting cell types, protoxylem and metaxylem, are patterned centripetally. We show that this patterning occurs through crosstalk between the vascular cylinder and the surrounding endodermis mediated by cell-to-cell movement of a transcription factor in one direction and microRNAs in the other. SHORT ROOT, produced in the vascular cylinder, moves into the endodermis to activate SCARECROW. Together these transcription factors activate MIR165a and MIR166b. Endodermally produced microRNA165/6 then acts to degrade its target mRNAs encoding class III homeodomain-leucine zipper transcription factors in the endodermis and stele periphery. The resulting differential distribution of target mRNA in the vascular cylinder determines xylem cell types in a dosage-dependent manner.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Comment in
-
Developmental biology: Roots respond to an inner calling.Nature. 2010 May 20;465(7296):299-300. doi: 10.1038/465299a. Nature. 2010. PMID: 20485422 No abstract available.
References
-
- Du TG, Schmid M, Jansen RP. Why cells move messages: The biological functions of mRNA localization. Semin Cell Dev Biol. 2007;18:171–177. - PubMed
-
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nature Genet. 2007;39:848–856. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

