Heme binding to the Mammalian circadian clock protein period 2 is nonspecific
- PMID: 20411915
- PMCID: PMC2873066
- DOI: 10.1021/bi901945w
Heme binding to the Mammalian circadian clock protein period 2 is nonspecific
Abstract
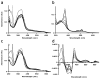
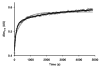
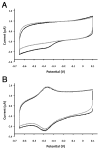
The mammalian circadian clock synchronizes physical and metabolic activity with the diurnal cycle through a transcriptional-posttranslational feedback loop. An additional feedback mechanism regulating clock timing has been proposed to involve oscillation in heme availability. Period 2 (PER2), an integral component in the negative feedback loop that establishes circadian rhythms in mammals, has been identified as a heme-binding protein. However, the majority of evidence for heme binding is based upon in vitro heme-binding assays. We sought to ascertain if these largely spectral assays could distinguish between specific and nonspecific heme interactions. Heme-binding properties by a number of other well-characterized proteins, all with no known biological role involving heme interaction, corresponded to those displayed by PER2. Site-directed mutants of putative heme-binding residues identified by MCD were unable to locate a specific heme-binding site on PER2. Protein film electrochemistry also indicates that heme binds PER2 nonspecifically on the protein surface. Our results establish the inability of qualitative in vitro assays to easily distinguish between specific and nonspecific heme binding. We conclude that heme binding to PER2 is likely to be nonspecific and does not involve the hydrophobic pocket within the PER2 PAS domains that in other PAS proteins commonly recognizes cofactors. These findings also question the significance of in vivo studies that implicate heme interactions with the clock proteins PER2 and nPAS2 in biological function.
Figures
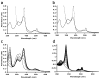
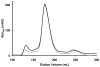
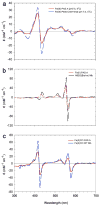

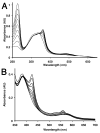
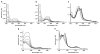
Similar articles
-
Heme-binding characteristics of the isolated PAS-B domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms.Biochim Biophys Acta. 2011 Feb;1814(2):326-33. doi: 10.1016/j.bbapap.2010.09.007. Epub 2010 Sep 29. Biochim Biophys Acta. 2011. PMID: 20887817
-
Early doors (Edo) mutant mouse reveals the importance of period 2 (PER2) PAS domain structure for circadian pacemaking.Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2756-61. doi: 10.1073/pnas.1517549113. Epub 2016 Feb 22. Proc Natl Acad Sci U S A. 2016. PMID: 26903623 Free PMC article.
-
Heme binding to human CLOCK affects interactions with the E-box.Proc Natl Acad Sci U S A. 2019 Oct 1;116(40):19911-19916. doi: 10.1073/pnas.1905216116. Epub 2019 Sep 16. Proc Natl Acad Sci U S A. 2019. PMID: 31527239 Free PMC article.
-
Development of circadian rhythms in mammalian systems.Biochem J. 2024 Dec 23;481(24):1967-1976. doi: 10.1042/BCJ20210060. Biochem J. 2024. PMID: 39714414 Review.
-
Genetics and neurobiology of circadian clocks in mammals.Cold Spring Harb Symp Quant Biol. 2007;72:251-259. doi: 10.1101/sqb.2007.72.052. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419282 Free PMC article. Review.
Cited by
-
Regulation of protein function and degradation by heme, heme responsive motifs, and CO.Crit Rev Biochem Mol Biol. 2022 Feb;57(1):16-47. doi: 10.1080/10409238.2021.1961674. Epub 2021 Sep 13. Crit Rev Biochem Mol Biol. 2022. PMID: 34517731 Free PMC article. Review.
-
Origin and functional diversification of PAS domain, a ubiquitous intracellular sensor.Sci Adv. 2023 Sep;9(35):eadi4517. doi: 10.1126/sciadv.adi4517. Epub 2023 Aug 30. Sci Adv. 2023. PMID: 37647406 Free PMC article.
-
An Analysis of the Multifaceted Roles of Heme in the Pathogenesis of Cancer and Related Diseases.Cancers (Basel). 2021 Aug 17;13(16):4142. doi: 10.3390/cancers13164142. Cancers (Basel). 2021. PMID: 34439295 Free PMC article. Review.
-
Heme binding properties of glyceraldehyde-3-phosphate dehydrogenase.Biochemistry. 2012 Oct 30;51(43):8514-29. doi: 10.1021/bi300863a. Epub 2012 Oct 15. Biochemistry. 2012. PMID: 22957700 Free PMC article.
-
Understanding the Logistics for the Distribution of Heme in Cells.JACS Au. 2021 Aug 10;1(10):1541-1555. doi: 10.1021/jacsau.1c00288. eCollection 2021 Oct 25. JACS Au. 2021. PMID: 34723258 Free PMC article. Review.
References
-
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. - PubMed
-
- Harms E, Kivimae S, Young MW, Saez L. Posttranscriptional and posttranslational regulation of clock genes. Journal of Biological Rhythms. 2004;19:361–373. - PubMed
-
- Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nature Reviews Genetics. 2001;2:702–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

