MicroRNA regulatory networks in cardiovascular development
- PMID: 20412767
- PMCID: PMC2922691
- DOI: 10.1016/j.devcel.2010.03.010
MicroRNA regulatory networks in cardiovascular development
Abstract
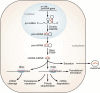
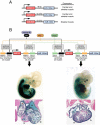
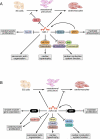
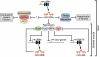
The heart, more than any other organ, requires precise functionality on a second-to-second basis throughout the lifespan of the organism. Even subtle perturbations in cardiac structure or function have catastrophic consequences, resulting in lethal forms of congenital and adult heart disease. Such intolerance of the heart to variability necessitates especially robust regulatory mechanisms to govern cardiac gene expression. Recent studies have revealed central roles for microRNAs (miRNAs) as governors of gene expression during cardiovascular development and disease. The integration of miRNAs into the genetic circuitry of the heart provides a rich and robust array of regulatory interactions to control cardiac gene expression. miRNA regulatory networks also offer opportunities for therapeutically modulating cardiac function through the manipulation of pathogenic and protective miRNAs. We discuss the roles of miRNAs as regulators of cardiac form and function, unresolved questions in the field, and issues for the future.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

