A pathobiont of the microbiota balances host colonization and intestinal inflammation
- PMID: 20413095
- PMCID: PMC2859213
- DOI: 10.1016/j.chom.2010.03.004
A pathobiont of the microbiota balances host colonization and intestinal inflammation
Abstract
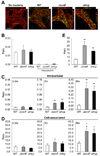
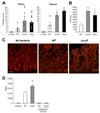
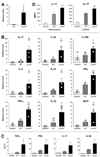
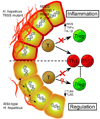
The gastrointestinal tract harbors a diverse microbiota that has coevolved with mammalian hosts. Though most associations are symbiotic or commensal, some resident bacteria (termed pathobionts) have the potential to cause disease. Bacterial type VI secretion systems (T6SSs) are one mechanism for forging host-microbial interactions. Here we reveal a protective role for the T6SS of Helicobacter hepaticus, a Gram-negative bacterium of the intestinal microbiota. H. hepaticus mutants with a defective T6SS display increased numbers within intestinal epithelial cells (IECs) and during intestinal colonization. Remarkably, the T6SS directs an anti-inflammatory gene expression profile in IECs, and CD4+ T cells from mice colonized with T6SS mutants produce increased interleukin-17 in response to IECs presenting H. hepaticus antigens. Thus, the H. hepaticus T6SS limits colonization and intestinal inflammation, promoting a balanced relationship with the host. We propose that disruption of such balances contributes to human disorders such as inflammatory bowel disease and colon cancer.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. - PubMed
-
- Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. - PubMed
-
- Das S, Chakrabortty A, Banerjee R, Chaudhuri K. Involvement of in vivo induced icmF gene of Vibrio cholerae in motility, adherence to epithelial cells, and conjugation frequency. Biochem Biophys Res Commun. 2002;295:922–928. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

