The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression
- PMID: 20413507
- PMCID: PMC2889767
- DOI: 10.2337/db09-0986
The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression
Abstract
Objective: Pancreatic islets of perinatal mice lacking the transcription factor Rfx3 exhibit a marked reduction in insulin-producing beta-cells. The objective of this work was to unravel the cellular and molecular mechanisms underlying this deficiency.
Research design and methods: Immunofluorescence studies and quantitative RT-PCR experiments were used to study the emergence of insulin-positive cells, the expression of transcription factors implicated in the differentiation of beta-cells from endocrine progenitors, and the expression of mature beta-cell markers during development in Rfx3(-/-) and pancreas-specific Rfx3-knockout mice. RNA interference experiments were performed to document the consequences of downregulating Rfx3 expression in Min6 beta-cells. Quantitative chromatin immunoprecipitation (ChIP), ChIP sequencing, and bandshift experiments were used to identify Rfx3 target genes.
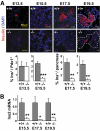
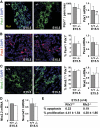
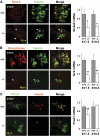
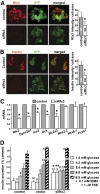
Results: Reduced development of insulin-positive cells in Rfx3(-/-) mice was not due to deficiencies in endocrine progenitors or beta-lineage specification, but reflected the accumulation of insulin-positive beta-cell precursors and defective beta-cells exhibiting reduced insulin, Glut-2, and Gck expression. Similar incompletely differentiated beta-cells developed in pancreas-specific Rfx3-deficient embryos. Defective beta-cells lacking Glut-2 and Gck expression dominate in Rfx3-deficent adults, leading to glucose intolerance. Attenuated Glut-2 and glucokinase expression, and impaired glucose-stimulated insulin secretion, were also induced by RNA interference-mediated inhibition of Rfx3 expression in Min6 cells. Finally, Rfx3 was found to bind in Min6 cells and human islets to two well-known regulatory sequences, Pal-1 and Pal-2, in the neuroendocrine promoter of the glucokinase gene.
Conclusions: Our results show that Rfx3 is required for the differentiation and function of mature beta-cells and regulates the beta-cell promoter of the glucokinase gene.
Figures



References
-
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A: Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell 2007;12:457–465 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

