Defining criteria for oligomannose immunogens for HIV using icosahedral virus capsid scaffolds
- PMID: 20416507
- PMCID: PMC2867452
- DOI: 10.1016/j.chembiol.2010.03.012
Defining criteria for oligomannose immunogens for HIV using icosahedral virus capsid scaffolds
Abstract
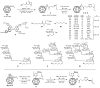
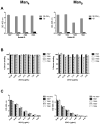
The broadly neutralizing antibody 2G12 recognizes a conserved cluster of high-mannose glycans on the surface envelope spike of HIV, suggesting that the "glycan shield" defense of the virus can be breached and may, under the right circumstances, serve as a vaccine target. In an attempt to recreate features of the glycan shield semisynthetically, oligomannosides were coupled to surface lysines on the icosahedral capsids of bacteriophage Q beta and cowpea mosaic virus (CPMV). The Q beta glycoconjugates, but not CPMV, presented oligomannose clusters that bind the antibody 2G12 with high affinity. However, antibodies against these 2G12 epitopes were not detected in immunized rabbits. Rather, alternative oligomannose epitopes on the conjugates were immunodominant and elicited high titers of anti-mannose antibodies that do not crossreact with the HIV envelope. The results presented reveal important design considerations for a carbohydrate-based vaccine component for HIV.
(c) 2010 Elsevier Ltd. All rights reserved.
Figures





References
-
- Astronomo RD, Lee HK, Scanlan CN, Pantophlet R, Huang CY, Wilson IA, Blixt O, Dwek RA, Wong CH, Burton DR. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J Virol. 2008;82:6359–6368. - PMC - PubMed
-
- Binley J. Specificities of broadly neutralizing anti-HIV-1 sera. Curr Opin HIV AIDS. 2009;4:364–372. - PubMed
-
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

