Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA
- PMID: 20418421
- PMCID: PMC2893475
- DOI: 10.1128/AEM.00436-10
Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA
Abstract
Previous studies identified the oleABCD genes involved in head-to-head olefinic hydrocarbon biosynthesis. The present study more fully defined the OleABCD protein families within the thiolase, alpha/beta-hydrolase, AMP-dependent ligase/synthase, and short-chain dehydrogenase superfamilies, respectively. Only 0.1 to 1% of each superfamily represents likely Ole proteins. Sequence analysis based on structural alignments and gene context was used to identify highly likely ole genes. Selected microorganisms from the phyla Verucomicrobia, Planctomyces, Chloroflexi, Proteobacteria, and Actinobacteria were tested experimentally and shown to produce long-chain olefinic hydrocarbons. However, different species from the same genera sometimes lack the ole genes and fail to produce olefinic hydrocarbons. Overall, only 1.9% of 3,558 genomes analyzed showed clear evidence for containing ole genes. The type of olefins produced by different bacteria differed greatly with respect to the number of carbon-carbon double bonds. The greatest number of organisms surveyed biosynthesized a single long-chain olefin, 3,6,9,12,15,19,22,25,28-hentriacontanonaene, that contains nine double bonds. Xanthomonas campestris produced the greatest number of distinct olefin products, 15 compounds ranging in length from C(28) to C(31) and containing one to three double bonds. The type of long-chain product formed was shown to be dependent on the oleA gene in experiments with Shewanella oneidensis MR-1 ole gene deletion mutants containing native or heterologous oleA genes expressed in trans. A strain deleted in oleABCD and containing oleA in trans produced only ketones. Based on these observations, it was proposed that OleA catalyzes a nondecarboxylative thiolytic condensation of fatty acyl chains to generate a beta-ketoacyl intermediate that can decarboxylate spontaneously to generate ketones.
Figures



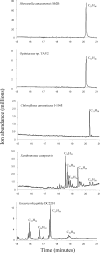
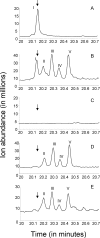
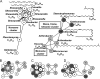
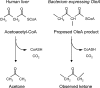
References
-
- Albro, P. W., and J. C. Dittmer. 1969. Biochemistry of long-chain, nonisoprenoid hydrocarbons. I. Characterization of the hydrocarbons of Sarcina lutea and the isolation of possible intermediates of biosynthesis. Biochemistry 8:394-405. - PubMed
-
- Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain nonisoprenoid hydrocarbons. III. The metabolic relationship of long-chain fatty acids and hydrocarbons and other aspects of hydrocarbon metabolism in Sarcina lutea. Biochemistry 8:1913-1918. - PubMed
-
- Albro, P. W., and J. C. Dittmer. 1969. The biochemistry of long-chain, nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by a cell-free preparation of Sarcina lutea. Biochemistry 8:3317-3324. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

