Application of multilocus sequence typing to study the genetic structure of megaplasmids in medicago-nodulating rhizobia
- PMID: 20418433
- PMCID: PMC2893487
- DOI: 10.1128/AEM.00251-10
Application of multilocus sequence typing to study the genetic structure of megaplasmids in medicago-nodulating rhizobia
Abstract
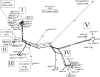
A multilocus sequence typing (MLST) analysis was used to examine the genetic structure and diversity within the two large extrachromosomal replicons in Medicago-nodulating rhizobia (Sinorhizobium meliloti and Sinorhizobium medicae). The allelic diversity within these replicons was high compared to the reported diversity within the corresponding chromosomes of the same strains (P. van Berkum et al., J. Bacteriol. 188:5570-5577, 2006). Also, there was strong localized linkage disequilibrium (LD) between certain pSymA loci: e.g., nodC and nifD. Although both of these observations could be explained by positive (or diversifying) selection by plant hosts, results of tests for positive selection did not provide consistent support for this hypothesis. The strong LD observed between the nodC and nifD genes could also be explained by their close proximity on the pSymA replicon. Evidence was obtained that some nodC alleles had a history of intragenic recombination, while other alleles of this locus had a history of intergenic recombination. Both types of recombination were associated with a decline in symbiotic competence with Medicago sativa as the host plant. The combined observations of LD between the nodC and nifD genes and intragenic recombination within one of these loci indicate that the symbiotic gene region on the pSymA plasmid has evolved as a clonal segment, which has been laterally transferred within the natural populations.
Figures



References
-
- Agapow, P.-M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1:101-102.
-
- Bailly, X., I. Olivieri, S. De Mita, J.-C. Cleyet-Marel, and G. Béna. 2006. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Mol. Ecol. 15:2719-2734. - PubMed
-
- Barran, L. R., E. S. P. Bromfield, and D. C. W. Brown. 2002. Identification and cloning of the bacterial nodulation specificity gene in the Sinorhizobium meliloti-Medicago laciniata symbiosis. Can. J. Microbiol. 48:765-771. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

